This project is done by me, aminrashidbeigi and Bita Jalali.
In this project, we explore a theoretical paper published in Neurips2017 Conference titled Learning Mixture of Gaussians with Streaming Data by doing some experimental tests on the performance of the algorithm. You can find the original paper here.
The authors of the paper proposed an algorithm with a theoretical guarantee for the problem of clustering the streaming data points where data points come from a Mixture of Gaussian Distribution. In particular, they showed that with high probability, the algorithm eventually finds (/learns) the mixture of Gaussians accurately. Moreover, they showed that the learning rate is near-optimal. Meaning that in each step, the error decreases optimally. Note that, as the problem of learning the mixture of Gaussian is NP-Hard, they used a simplification assumption for the data to make the problem tractable. The simplification assumption they used is that data comes from a mixture of well-separated Gaussians. (i.e. the centers of any two clusters are far enough so that the algorithm can decide computationally easily whether the point belongs to the first cluster or the second one.)
Although the assumption allows them to have a good performance for this problem, one question might arise:
Is the analysis of the paper tight?
One way to find an answer to this question is to test the algorithm's performance by running some experimental tests. Of course, the experimental tests just provide evidence and they are not proof.
In the following, we will first give a bit explanation about the general setup and data distribution in the first section. Then we explain the theorems about the performance of the algorithm. Finally, we test the algorithm performance by generating data from the mixture of Gaussian Distribution and run our implementation of the paper's algorithm
Data is drawn from a mixture of a mixture of k spherical Gaussians distributions,
i.e. for all
where
For simplicity, we assumed all
As we stated earlier, the paper used a simplification assumption for the data. In particular, any two centers
We will explain more about the constant C in the next sections.
100 points drawn from the distribution are depicted here. (note that we have shown a 2-d subspace from the original d-dimensional space of data points.
You can see there are k clusters.
temp = generator.generate_samples(100)
plt.plot(temp[0, :], temp[1,:], 'x')
plt.title("Space Overview")
plt.show()The algorithm consists of two parts:
1 . Algorithm1 (initialization algorithm)
2 . Algorithm2: (a) Streaming using Lloyd's Heuristic, (b) Expectation Maximization (i.e.Soft version of Lloyd's Heuristic)
Note that Expectation Maximization part is available in another file named "EM".
In the initialization part, we try to find k initial centers for k clusters. To do so, we first use SVD-style operations to project d dimensional data points into a k-dimensional space. These operations project will de-noise the data, making it easy for the algorithm to identify all k distinct clusters from few data points. After we find k clusters centers in the projected space, we project them to the original d-dimensional space. This way, we have a good initialization for the k centers.
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import ortho_group
from scipy import linalg
import random
from sklearn import neighbors
import sklearn.metrics.pairwise
from scipy.sparse import csgraph
import random
import pandas as pd
d = 10
k = 5
scaler = 10000
sigma = 100
num_of_samples = int(d*d*d*k*k*k*np.log(d*k)) #N0
w = np.ones(k) / k
## For initializatoin, we need C to be at least OMEGA((klogk)^1/4). So:
C = 10*((k*np.log(k))**(1/4))
C2 = (np.log(k)**(1/2))
##TODO: Change max to min but make sure that you ignore the diagonal values as they are zeros and they should be ignored.
def generate_distant_means(dimension, C, std):
mu = np.random.rand(d, k) * scaler
dist = sklearn.metrics.pairwise.euclidean_distances(mu.T)
dist = dist + np.identity(k, dtype = float) * 2 * C * sigma
min_dist = np.min(dist)
while (min_dist < C * sigma):
print ("close_initial_points_occured, start again")
mu = np.random.rand(d, k) * scaler
dist = sklearn.metrics.pairwise.euclidean_distances(mu.T)
dist = dist + np.identity(k, dtype = float) * 2 * C * sigma
min_dist = np.min(dist)
return mu
mu = generate_distant_means(d, C, sigma)
print ("True Centers for Clusters")
tmp = pd.DataFrame(mu)
tmpTrue Centers for Clusters
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| 0 | 1006.573560 | 7457.903141 | 8607.361205 | 5805.062752 | 159.473022 |
| 1 | 7981.089236 | 4519.639663 | 8843.143067 | 7082.612751 | 1638.453342 |
| 2 | 1786.589980 | 7512.222316 | 4686.177980 | 1062.615207 | 6604.511910 |
| 3 | 3298.659779 | 8089.362560 | 6149.617684 | 4718.901034 | 7559.245778 |
| 4 | 8864.726130 | 5712.461236 | 1803.188495 | 7849.071028 | 7154.246706 |
| 5 | 7637.138501 | 6373.934348 | 3263.415539 | 1354.105588 | 2977.484326 |
| 6 | 6208.651794 | 2407.470493 | 1071.618747 | 5643.999592 | 3806.159423 |
| 7 | 2173.689721 | 828.673876 | 1893.492336 | 2113.814978 | 2271.450202 |
| 8 | 5882.354979 | 1509.902750 | 6242.963681 | 8404.339237 | 9478.986709 |
| 9 | 9055.694116 | 9003.731212 | 6530.270935 | 1714.366744 | 2276.888733 |
Sample Generator Class
class SampleGenerator:
def __init__(self, weights, mu, sigma, d):
self.weights = weights
self.mu = mu
self.sigma=sigma
self.d = d
def draw_a_sample(self):
rand = random.uniform(0,1)
for i in range(len(self.weights)):
rand -= self.weights[i]
if (rand <=0):
return np.random.multivariate_normal(self.mu[:,i], self.sigma * np.identity(self.d), 1).T
def generate_samples(self, sample_size):
samples = []
for i in range(sample_size):
if len(samples) == 0:
samples = self.draw_a_sample()
else:
samples = np.append(samples, self.draw_a_sample(), axis=1)
return samples
generator = SampleGenerator(w, mu, sigma, d)In this part, we are going to find a k dimensional space to reduce the noise of each cluster. This way, all points from a single clusters will become closer to each other, allowing us to use Nearest Neighbor Graph to identify each cluster easily.
N0 = num_of_samples
S = np.zeros((d, d))
k_2 = int(10 * k * np.log(k))
B = int(d * np.log(d))
U = ortho_group.rvs(dim=d)[:, :k] # Choosing k orthonormal vectors in Rd space
for t in range(N0 - k_2):
if t % B == B - 1:
Q, R = linalg.qr(np.matmul(S, U))
U = Q
S = np.zeros((d, d))
reshaped_sample = generator.draw_a_sample().reshape(-1, 1)
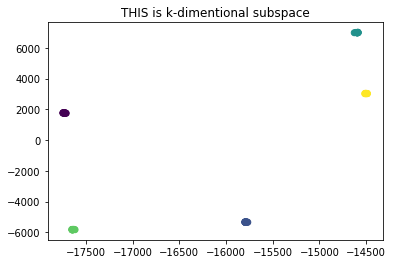
S = S + np.matmul(reshaped_sample, reshaped_sample.T)First, we project points to the k-dimensional space to reduce the noise, then using each point as a node, we build a nearest neighbor graph. Each connected component in the graph would represent a cluster.
X0 = generator.generate_samples(k_2)
save_U = U
U = U[:, 0:k]
components = np.matmul(U.T, X0)
adj = neighbors.kneighbors_graph(np.transpose(components), 3)
n_components, labels = csgraph.connected_components(adj)
fig, ax = plt.subplots()
ax.scatter(components[0], components[1], c=labels)
plt.title("THIS is k-dimentional subspace")
plt.show()components = np.matmul(U, components)
fig, ax = plt.subplots()
ax.scatter(components[0], components[1], c=labels)
plt.title("This is Original Space")
plt.show()components_mean = np.zeros((k, d))
components_by_labels = []
for c in range(k):
list_of_indices = np.where(labels == c)[0]
components_mean[c] = np.mean(components[:, list_of_indices], axis=1)
print("Components mean:")
pd.DataFrame(components_mean.T)Components mean:
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| 0 | 1004.659735 | 8602.688447 | 156.998482 | 7457.114878 | 5810.833626 |
| 1 | 7981.449254 | 8842.145440 | 1632.891263 | 4523.029546 | 7070.616973 |
| 2 | 1790.712167 | 4682.362683 | 6602.861160 | 7512.045539 | 1061.278888 |
| 3 | 3296.125969 | 6153.697604 | 7558.724763 | 8092.409179 | 4712.346784 |
| 4 | 8863.349343 | 1806.231879 | 7152.189040 | 5715.178904 | 7845.894733 |
| 5 | 7628.245490 | 3258.963132 | 2980.991991 | 6375.568134 | 1361.207542 |
| 6 | 6215.085481 | 1067.861391 | 3807.590765 | 2404.595059 | 5642.911509 |
| 7 | 2169.924317 | 1889.896869 | 2275.113901 | 832.385092 | 2119.977969 |
| 8 | 5885.804747 | 6239.186480 | 9477.207032 | 1509.366177 | 8414.359976 |
| 9 | 9054.406525 | 6531.609386 | 2277.226207 | 9008.111500 | 1717.347722 |
print ("TRUE centers")
pd.DataFrame(mu)TRUE centers
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| 0 | 1006.573560 | 7457.903141 | 8607.361205 | 5805.062752 | 159.473022 |
| 1 | 7981.089236 | 4519.639663 | 8843.143067 | 7082.612751 | 1638.453342 |
| 2 | 1786.589980 | 7512.222316 | 4686.177980 | 1062.615207 | 6604.511910 |
| 3 | 3298.659779 | 8089.362560 | 6149.617684 | 4718.901034 | 7559.245778 |
| 4 | 8864.726130 | 5712.461236 | 1803.188495 | 7849.071028 | 7154.246706 |
| 5 | 7637.138501 | 6373.934348 | 3263.415539 | 1354.105588 | 2977.484326 |
| 6 | 6208.651794 | 2407.470493 | 1071.618747 | 5643.999592 | 3806.159423 |
| 7 | 2173.689721 | 828.673876 | 1893.492336 | 2113.814978 | 2271.450202 |
| 8 | 5882.354979 | 1509.902750 | 6242.963681 | 8404.339237 | 9478.986709 |
| 9 | 9055.694116 | 9003.731212 | 6530.270935 | 1714.366744 | 2276.888733 |
As we know in the Theorem 3 of the paper,
if each two centers have enough seperations,
then with high probability
def distance_calculator(mu, mu_t):
comparision_matrix = sklearn.metrics.pairwise.euclidean_distances(mu.T, mu_t.T)
distance_vector = np.min(comparision_matrix, axis=0)
distance_sum = np.sum(np.min(comparision_matrix, axis=0))
return distance_sum, distance_vectordistance_sum, distance_vector = distance_calculator(mu, components_mean.T)
DF = pd.DataFrame(distance_vector)
DF.columns = ["Distance"]
print ("Distance between each cluster center and algorithm's output for that center:")
print(DF)
print("Sum of distances:")
print(distance_sum)Distance between each cluster center and algorithm's output for that center:
Distance
0 13.315987
1 11.234860
2 8.680253
3 8.543820
4 20.759095
Sum of distances:
62.53401504530203
theorem3_bound= C*sigma/20
print ("Theorem 3 Bound: C/20 * sigma")
theorem3_boundTheorem 3 Bound: C/20 * sigma
8.421337295817725
As you can see, the TRUE MEANS are very close to initial algorithm's output as desired.
In this part, we are going to use the Lloyd Heuristic for clustering. Uppon the arrival of each data point, we firstly assign it to its closest center and then update that center.
from numpy import linalg as LA
import copy
mu01 = components_mean.T
def streaming_kmeans(n, input_mu, num_of_logs):
mu0 = copy.deepcopy(input_mu)
distance_log = []
eta = 3 * k * (np.log2(3 * n)) / n
cut = int(n / num_of_logs)
for t in range(n):
if t % cut == 0:
d, _ = distance_calculator(mu, mu0)
distance_log.append(d)
x = generator.draw_a_sample()
dis = LA.norm((x - mu0), axis=0)
i = np.argmin(dis)
x = x.reshape(-1)
mu0[:, i] = np.multiply((1 - eta), mu0[:, i]) + np.multiply(eta, x)
return mu0, distance_logIntuitively, this theorem upper bounds the error when we have seen enough data.
Let
In the following, we will verify theorem 1 experimentally.
def theorem_1(mu, N, sigma, K, C, d):
return (np.max(LA.norm(mu,axis=0)) / N) + (K**3 * (sigma**2 * d * np.log(N) / N) + np.exp(-C**2 / 8) * (C**2 + K)* sigma**2)num_of_logs = 1000
run_count = 10
error_matrix = np.zeros((run_count, num_of_logs))
Ns = []
n = 2*int(d*d*d*k*k*k*np.log(d*k))
for run in range(run_count):
mu_t, distance_log = streaming_kmeans(n, mu01, num_of_logs)
error_matrix[run] = distance_log[:num_of_logs]
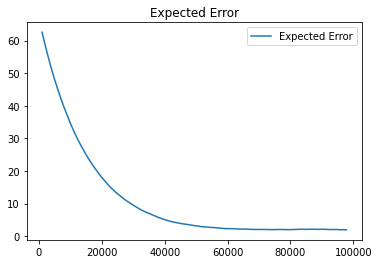
df=pd.DataFrame(error_matrix)mean_error = np.mean(error_matrix, axis=0)
diagram_cut = 100
theorem_1_log = []
Ns = []
for i in range(num_of_logs):
cut = int(n / num_of_logs)
theorem_1_log.append(theorem_1(mu, cut * (i+1), sigma, k, C, d))
Ns.append(cut * (i+1))
p1, =plt.plot(Ns[:diagram_cut], mean_error[:diagram_cut], label='Expected Error')
plt.legend(loc='best')
plt.title('Expected Error')
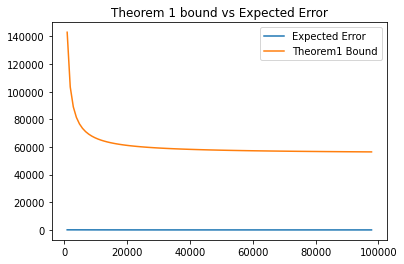
plt.show()p1, =plt.plot(Ns[:diagram_cut], mean_error[:diagram_cut], label='Expected Error')
p2, =plt.plot(Ns[:diagram_cut], theorem_1_log[:diagram_cut], label='Theorem1 Bound')
plt.legend(loc='best')
plt.title('Theorem 1 bound vs Expected Error')
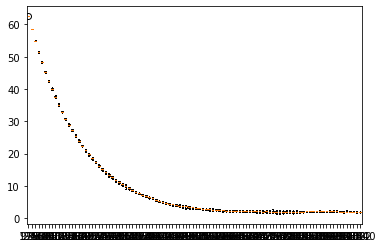
plt.show()boxplot_dict = {}
for i in range(diagram_cut):
boxplot_dict[cut * (i+1)] = [np.mean(error_matrix[:, i]), np.min(error_matrix[:, i]), np.max(error_matrix[:, i]), ]
fig, ax = plt.subplots()
ax.boxplot(boxplot_dict.values())
ax.set_xticklabels(boxplot_dict.keys())
plt.plot()[]
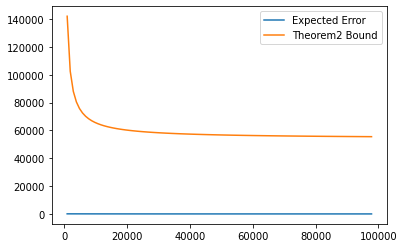
The streaming part of the algorithm works even when the data seperation C is much less. (when $ C \geq \Omega (\log k)^{1/2} $)
In this part, we will verify it experimentally in the following cell.
theorem_2_log = []
for i in range(num_of_logs):
cut = int(n / num_of_logs)
theorem_2_log.append(theorem_1(mu, cut * (i+1), sigma, k, C2, d))
p1, =plt.plot(Ns[:diagram_cut], mean_error[:diagram_cut], label='Expected Error')
p2, =plt.plot(Ns[:diagram_cut], theorem_2_log[:diagram_cut], label='Theorem2 Bound')
plt.legend(loc='best')
plt.show()