Matching single cells across modalities with contrastive learning and optimal transport.
Preprint available on bioRxiv (Gossi et al., 2022)
-
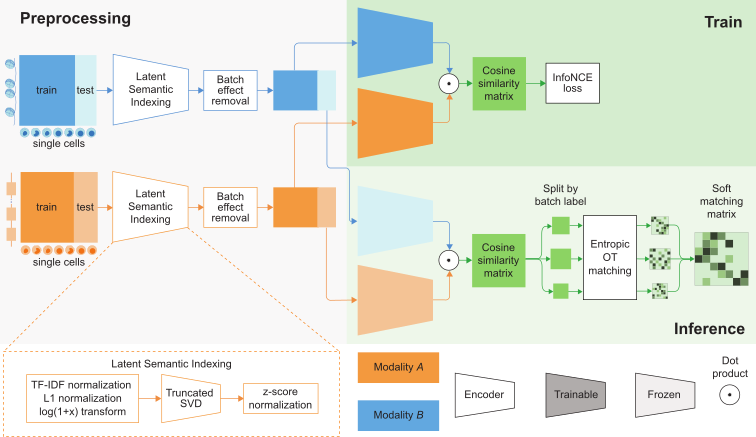
MatchCLOT is a computational framework that is able to match single-cells measured using different omic modalities.
-
With paired multi-omic data, MatchCLOT uses contrastive learning to learn a common representation between two modalities after applying a preprocessing pipeline for normalization and dimensionality reduction.
-
Based on the similarities between the cells in the learned representation, MatchCLOT finds a matching between the cell profiles in the two omic modalities using entropic optimal transport.
-
Pretrained MatchCLOT models can be applied to new unpaired multiomic data to match two modalities at single-cell resolution.
NOTE: The package and the documentation are under development.
MatchCLOT is available on PyPI and can be installed with pip:
pip install matchclot
The documentation is available at https://matchclot.readthedocs.io/en/latest/