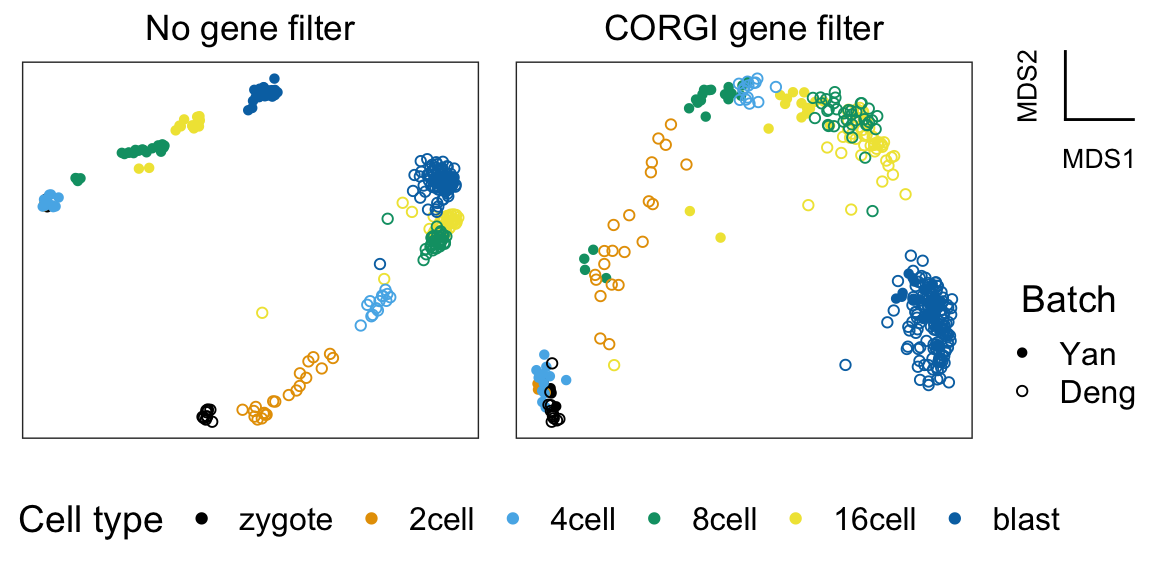
CORGI (Curve-fit Objective Ranking of Gene Importance) is a gene filter for integrating two single-cell RNAseq trajectory datasets. Conceptually, CORGI selects genes that highlight the shared heterogenity in both datasets while rejecting genes that contributes to batch effects.
For example, MDS run on genes selected by CORGI can remove the "batch effect" between mouse and human pre-implantation embryogenesis:
In R, run
install.packages("devtools")
devtools::install_github("YutongWangUMich/corgi")
For a quick introduction, check out the vignette: integrating pre-implantation embryogenesis between mouse and human.