TIMSCONVERT is a workflow designed for mass spectrometrists that allows for the conversion of raw data from Bruker timsTOF Pro and fleX mass spectrometers (i.e. .d directory containing BAF, TDF, and TSF files) to open source data formats (i.e. mzML and imzML) that:
- are compatible with downstream open source data analysis platorms.
- incorporate trapped ion mobility spectrometry (TIMS) data into these open source formats.
- do not require any programming experience.
If you use TIMSCONVERT, please cite us:
Gordon T. Luu, Michael A. Freitas, Itzel Lizama-Chamu, Catherine S. McCaughey, Laura M. Sanchez, Mingxun Wang. (2022). TIMSCONVERT: A workflow to convert trapped ion mobility data to open formats. Bioinformatics; btac419. DOI: 10.1093/bioinformatics/btac419.
- LC-MS(/MS) (BAF) → mzML
- LC-TIMS-MS(/MS) (TDF) → mzML
- MALDI-MS(/MS) Dried Droplet (TSF) → mzML
- MALDI-MS Imaging Mass Spectrometry (TSF) → imzML
- MALDI-TIMS-MS(/MS) Dried Droplet (TDF) → mzML
- MALDI-TIMS-MS Imaging Mass Spectrometry (TDF) → imzML (does not incorporate ion mobility data)
Please note that TIMSCONVERT is still actively under development and new changes are being pushed regularly.
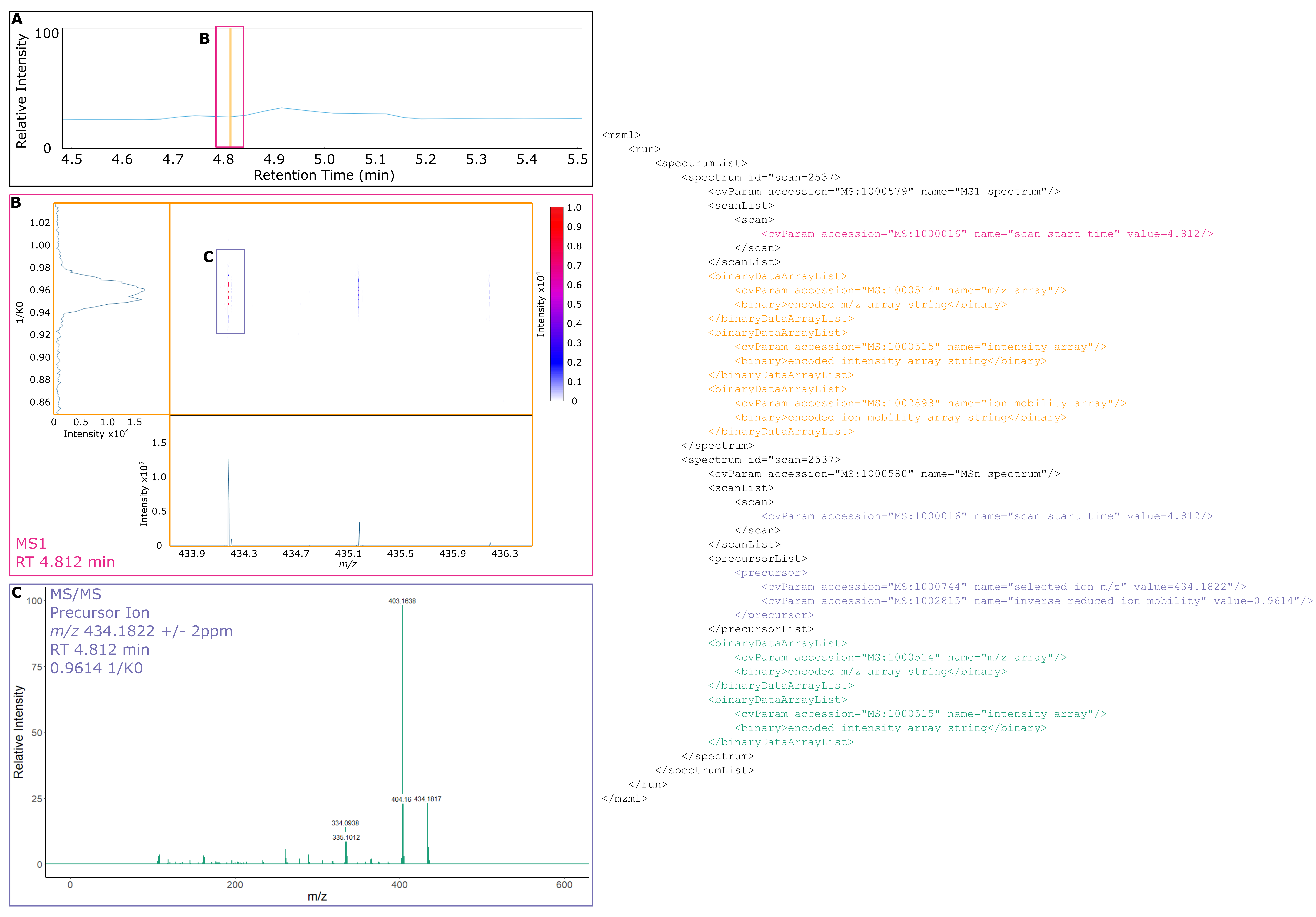
Example of dimensionality of LC-TIMS-MS/MS data and a simplified corresponding mzML schema. Elements in the simplified mzML schema are color coded by the corresponding data in the chromatograms/spectra. At a given retention time in the chromatogram (indicated in pink), an MS1 spectrum may be visualized in the form of a three dimensional plot generated from m/z, intensity, and ion mobility arrays (indicated in orange). Precursor ions of interest found in MS1 spectra (indicated in purple) can be further analyzed by plotting m/z and intensity arrays for MS/MS spectra (indicated in teal). These spectral identifiers (i.e. retention time, precursor m/z, precursor 1/K0) may be used to locate the corresponding MS/MS spectrum from Bruker DataAnalysis or other data visualization software of choice.
The web version of TIMSCONVERT currently supports various data formats listed below. No installation is necessary. All that's required is a GNPS account.
- Upload your data to GNPS (instructions).
- Go to the TIMSCONVERT workflow page.
- Select your dataset.
- Submit your run.
- You will receive an email when your job has completed.
| Acquisition Mode | Raw File Format | Converted File Format | Compatible? |
|---|---|---|---|
| LC-MS(/MS) | .d/BAF | mzML | ❌ |
| LC-TIMS-MS(/MS) | .d/TDF | mzML | ✔️ |
| MALDI-MS(/MS) Dried Droplet | .d/TSF | mzML | ✔️ |
| MALDI-MS Imaging Mass Spectrometry | .d/TSF | imzML | ✔️ |
| MALDI-TIMS-MS(/MS) Dried Droplet | .d/TDF | mzML | ✔️ |
| MALDI-TIMS-MS Imaging Mass Spectrometry | .d/TDF | imzML | ✔️ |
If you prefer to run TIMSCONVERT locally via Nextflow or the CLI, you can set up an environment to do so. Please note that TIMSCONVERT should be run under Linux or Windows. macOS is not supported.
- Download and install Anaconda for Linux. Follow the prompts to complete installation. Anaconda3-2021.11 for Linux is used as an example here.
wget https://repo.anaconda.com/archive/Anaconda3-2021.11-Linux-x86_64.sh
bash /path/to/Anaconda3-2021.11-Linux-x86_64.sh
- Add
anaconda3/binto PATH.
export PATH=$PATH:/path/to/anaconda3/bin
- Download and install Anaconda for Windows. Follow the prompts to complete installation.
- Run
Anaconda Prompt (R-MINI~1)as Administrator.
- Create a conda instance. You must be using Python 3.7. Newer versions of Python are not guaranteed to be compatible with Bruker's API in Linux.
conda create -n timsconvert python=3.7
- Activate conda environment.
conda activate timsconvert
- (Optional, for Nextflow workflow only) Install Nextflow.
conda install -c bioconda nextflow
- Download TIMSCONVERT by cloning the Github repo (you will need to have Git and
ensure that the option to enable symbolic links was checked during installation). It may be necessary to explicitly
allow for the use of symbolic links by adding the
-c core.symlinks=trueparameter on Windows.
git clone https://www.github.com/gtluu/timsconvert
or
git clone -c core.symlinks=true https://www.github.com/gtluu/timsconvert
- Install dependencies.
# TIMSCONVERT dependencies
pip install -r /path/to/timsconvert/requirements.txt
- You will also need to install our forked version of pyimzML, which has added support for ion mobility arrays in imzML data from imaging mass spectrometry experiments.
pip install git+https://github.com/gtluu/pyimzML
A Nextflow workflow has been provided to run TIMSCONVERT.
- Run the
nextflow.nfscript provided in this repo and specify your input directory and experiment type. Unless specified, all other default parameters for all other values will be used. See below for an explanation of all parameters.
nextflow run /path/to/timsconvert/nextflow.nf --input /path/to/your/data
- Depending on the size of your data/number of files, TIMSCONVERT may take some time to finish conversion.
The CLI version of TIMSCONVERT supports conversion of all experimental data types specified above.
- Use
bin/run.pyto run TIMSCONVERT. The input directory and experiment type. Unless specified, all other default parameters for all other values will be used. See below for an explanation of all parameters.
python /path/to/timsconvert/bin/run.py --input /path/to/data
or
python3 /path/to/timsconvert/bin/run.py --input /path/to/data
- Depending on the size of your data/number of files, TIMSCONVERT may take some time to finish conversion.
A Dockerfile has also been provided to run TIMSCONVERT inside a Docker container.
- Build the Docker image.
docker build --tag timsconvert -f /path/to/timsconvert/Dockerfile .
- Run the Docker image in a container.
docker run --rm -it -v /path/to/data:/data timsconvert --input /data --outdir /data
Required Parameters
--input Bruker .d file containing TSF/TDF or directory containing multiple Bruker .d files.
Optional Parameters
--outdir Path to folder in which to write output file(s). Defaults to .d source folder.
--outfile User defined filename for output if converting a single file. If input is a folder
with msdultiple .d files, this parameter should not be used as it results in each file
overwriting the previous due to having the same filename.
--mode Choose whether to export spectra in "raw" or "centroid" formats. Deafults to
"centroid".
--compression Choose between ZLIB compression ("zlib") or no compression ("none"). Defaults to "zlib".
TIMSCONVERT Optional Parameters
--ms2_only Boolean flag that specifies only MS2 spectra should be converted.
--exclude_mobility Boolean flag used to exclude trapped ion mobility spectrometry data from exported data.
Precursor ion mobility information is still exported. Recommended when exporting in profile
mode due to file size.
--encoding Choose encoding for binary arrays. 32-bit ("32") or 64-bit ("64"). Defaults to 64-bit.
--barebones_metadata Only use basic mzML metadata. Use if downstream data analysis tools throw errors with
descriptive CV terms.
--profile_bins Number of bins used to bin data when converting in profile mode. A value of 0 indicates no
binning is performed. Default s to 0
--maldi_output_file For MALDI dried droplet data, whether individual scans should be placed in
individual files ("individual") or all into a single file ("combined").
Defaults to combined.
--maldi_plate_map Plate map to be used for parsing spots if --maldi_output_file == "individual".
Should be a .csv file with no header/index.
--imzml_mode Whether imzML files should be written in "processed" or "continuous" mode. Defaults
to "processed".
TIMSCONVERT System Parameters
--lcms_backend Choose whether to use "timsconvert" or "tdf2mzml" backend for LC-TIMS-MS/MS data conversion.
--chunk_size Relative size of chunks of spectral data that are parsed and subsequently written at
once.
--verbose Boolean flag to determine whether to print logging output.
tdf2mzml Optional Parameters
--start_frame Start frame.
--end_frame End frame.
--precision Precision.
--ms1_threshold Intensity threshold for MS1 data.
--ms2_threshold Intensity threshold for MS2 data.
--ms2_nlargest N Largest MS2.
Get test data
cd test
make download_test
Run workflow
make run_test
make run_nextflow_test
v 1.0.0
- Initial release.
- Compatibility for ESI and MALDI based experiments.