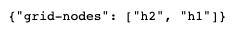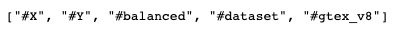A Federated Learning repository for simulating decentralized training for common biomedical use-cases
- About
- Installation and Initialization
- Local Execution
- Remote Execution
- Running DVC stages
- Notebooks
- Hyperparameter Optimization
- Testing
- Known Issues
- Tutorials / References
- Project Status
- Acknowledgements
- Quality information exist as islands on gadgets like cell phones and PCs over the globe and are protected by severe security safeguarding laws.
- Federated Learning gives an astute methods for associating AI models to these incoherent information paying little heed to their areas, and all the more significantly, without penetrating protection laws.
- In biomedical research, sharing and use of human biomedical data is also heavily restricted and regulated by multiple laws. Such data-sharing restrictions allow keeping privacy of the patients but at the same time it impedes the pace of biomedical research, slows down the development of treatments of various diseases and often costs human lives.
- COVID-19 pandemic is unfortunately a good illustration of how inaccessibility of clinical training data leads to casualties that can be otherwise avoided.
- This repository is devoted to addressing this issue for the most common biomedical use-cases, like gene expression data.
- It is an introductory project for simulating
easy-to-deployFederated Learning, for decentralized biomedical datasets.- A user can either simulate FL training locally (using
localhost), or remotely (onseveral machines). - A user can also compare centralized vs decentralized train metrics.
- A user can either simulate FL training locally (using
- Technology Stack used:
- Example Dataset used:
- GTEx: The Common Fund's Genotype-Tissue Expression (GTEx) Program established a data resource and tissue bank to study the relationship between genetic variants (inherited changes in DNA sequence) and gene expression (how genes are turned on and off) in multiple human tissues and across individuals.
- NOTE: All the testing has been done on a MacOS / Linux based system
- Step 1: Install Docker & Docker-Compose, and pull required images from DockerHub
- To install
Docker, just follow the docker documentation. - To install
Docker-Compose, just follow the docker-compose documentation. - Start your
docker daemon - Pull grid-node image :
docker pull srijanverma44/grid-node:v028 - Pull grid-network image :
docker pull srijanverma44/grid-network:v028
Size of grid-node ~= 2GB, and that of grid-network ~= 300MB. That is, image sizes are large!
- To install
- Step 2: Install dependencies via conda
- Install Miniconda, for your operating system, from https://conda.io/miniconda.html
git clone https://github.com/vermasrijan/srijan-gsoc-2020.gitcd srijan-gsoc-2020conda env create -f environment.ymlconda activate pysyft_v028(orsource activate pysyft_v028for older versions of conda)
- Step 3: Install GTEx
V8Dataset- Pull
samplesandexpressionsdata using the following command:
- Pull
dvc pull
- The above command will download GTEx
samples + expressionsdata insidedata/gtexdirectory, from Google Drive remote repository.- Initially, you may be prompted to enter a verification code, i.e., you'll have to give DVC an access to your Google Drive API.
- For that, go to the URL which may be displayed on your CLI, copy the code, paste it at CLI and press Enter. (For more info, refer 1 & 2)
src/initializer.pyis a python script for initializing either a centralized training, or a decentralized one.- This script will create a compose yaml file, initialize
client/networkcontainers, execute FL/centralized training and will finally stop running containers (for network/nodes).
- Make sure your
docker daemonis running - Run the following command -
python src/initializer.py
Usage: initializer.py [OPTIONS]
Options:
--samples_path TEXT Input path for samples
--expressions_path TEXT Input for expressions
--train_type TEXT Either centralized or decentralized fashion
--dataset_size INTEGER Size of data for training
--split_type TEXT balanced / unbalanced / iid / non_iid
--split_size FLOAT Train / Test Split
--n_epochs INTEGER No. of Epochs / Rounds
--metrics_path TEXT Path to save metrics
--model_save_path TEXT Path to save trained models
--metrics_file_name TEXT Custom name for metrics file
--no_of_clients INTEGER Clients / Nodes for decentralized training
--swarm TEXT Option for switching between docker compose vs docker stack
--no_cuda TEXT no_cuda = True means not to use CUDA. Default --> use CPU
--tags TEXT Give tags for the data, which is to be sent to the nodes
--node_start_port TEXT Start port No. for a node
--grid_address TEXT grid address for network
--grid_port TEXT grid port for network
--help Show this message and exit.
- Example command:
python src/initializer.py --train_type centralized --dataset_size 17000 --n_epochs 50
Centralized trainingexample output, using 50 epochs:
============================================================
----<DATA PREPROCESSING STARTED..>----
----<STARTED TRAINING IN A centralized FASHION..>----
DATASET SIZE: 17000
Epoch: 0 Training loss: 0.00010540 | Training Accuracy: 0.1666
Epoch: 1 Training loss: 0.00010540 | Training Accuracy: 0.1669
.
.
Epoch: 48 Training loss: 9.3619e-05 | Training Accuracy: 0.4356
Epoch: 49 Training loss: 9.3567e-05 | Training Accuracy: 0.4359
---<SAVING METRICS.....>----
============================================================
OVERALL RUNTIME: 43.217 seconds
dvc repro centralized_train
- Example command:
python src/initializer.py --train_type decentralized --dataset_size 17000 --n_epochs 50 --no_of_clients 2
Decentralized trainingexample output, using 50 epochs:
- Distributed information, like total no. of samples with each client, will be displayed first.
============================================================
----<DATA PREPROCESSING STARTED..>----
----<STARTED TRAINING IN A decentralized FASHION..>----
DATASET SIZE: 17000
TOTAL CLIENTS: 2
DATAPOINTS WITH EACH CLIENT:
client_h1: 8499 ; Label Count: {0: 1445, 1: 1438, 2: 1429, 3: 1432, 4: 1394, 5: 1361}
client_h2: 8499 ; Label Count: {0: 1388, 1: 1395, 2: 1404, 3: 1401, 4: 1439, 5: 1472}
---<STARTING DOCKER IMAGE>----
====DOCKER STARTED!=======
Go to the following addresses: ['http://0.0.0.0:5000', 'http://0.0.0.0:5000/connected-nodes', 'http://0.0.0.0:5000/search-available-tags', 'http://0.0.0.0:3000', 'http://0.0.0.0:3001']
Press Enter to continue...
-------<USING CPU FOR TRAINING>-------
WORKERS: ['h1', 'h2']
Train Epoch: 0 | With h2 data |: [8499/16998 (50%)] Train Loss: 0.000211 | Train Acc: 0.164
Train Epoch: 0 | With h1 data |: [16998/16998 (100%)] Train Loss: 0.000211 | Train Acc: 0.192
Train Epoch: 1 | With h2 data |: [8499/16998 (50%)] Train Loss: 0.000211 | Train Acc: 0.172
Train Epoch: 1 | With h1 data |: [16998/16998 (100%)] Train Loss: 0.000211 | Train Acc: 0.229
.
.
Train Epoch: 49 | With h2 data |: [8499/16998 (50%)] Train Loss: 0.000187 | Train Acc: 0.384
Train Epoch: 49 | With h1 data |: [16998/16998 (100%)] Train Loss: 0.000187 | Train Acc: 0.389
---<STOPPING DOCKER NODE/NETWORK CONTAINERS>----
381c4f79fb5c
c203c2f6fd62
1d3ccce7f732
---<SAVING METRICS.....>----
============================================================
OVERALL RUNTIME: 380.418 seconds
dvc repro decentralized_train
- NOTE: By default, metrics will be saved in
data/metricsdirectory. - You can pass in the
--metrics_path <path>flag to change the default directory.
- Following is what you may see at http://0.0.0.0:5000
- Following is what you may see at http://0.0.0.0:5000/connected-nodes
- Following is what you may see at http://0.0.0.0:5000/search-available-tags
- Following is what you may see at http://0.0.0.0:3000
- Make sure all Firewalls are disabled on both, client and server side.
- Docker-compose will be required in this section.
docker-compose -f gridnetwork-compose.yml up
- STEP 1: Configure the environment variable called
NETWORK, and replace it with <SERVER_IP_ADDRESS> - STEP 2:
docker-compose -f gridnode-compose.yml up. You can edit this compose file to add more clients, if you'd like.
- NOTE: Remote execution has not yet been tested properly.
- DVC stages are in
dvc.yamlfile, to run dvc stage just usedvc repro <stage_name>
- Notebooks, given in this repository, simulate decentralized training using 2 clients.
- Docker-compose will be required in this section as well!
- STEP 1:
docker-compose -f notebook-docker-compose.yml up - STEP 2:
conda activate pysyft_v028(orsource activate pysyft_v028for older versions of conda) - STEP 3: Go to the following addresses:
['http://0.0.0.0:5000', 'http://0.0.0.0:5000/connected-nodes', 'http://0.0.0.0:5000/search-available-tags', 'http://0.0.0.0:3000', 'http://0.0.0.0:3001']
- STEP 4: Initialize
jupyter lab - STEP 5: Run data owner notebook:
notebooks/data-owner_GTEx.ipynb - STEP 6: Run model owner notebook:
notebooks/model-owner_GTEx.ipynb - STEP 7: STOP Node/Network running containers:
docker rm $(docker stop $(docker ps -a -q --filter ancestor=srijanverma44/grid-network:v028 --format="{{.ID}}"))
docker rm $(docker stop $(docker ps -a -q --filter ancestor=srijanverma44/grid-node:v028 --format="{{.ID}}"))
NOTE:
- Notebooks given in this repository have been taken from this branch and have been modified.
python src/tune.py --help
- In Progress...
- Test Centralized training:
dvc repro centralized_test
- Test Decentralized training:
dvc repro decentralized_test
- While creating an environment:
- While creating an env. on a linux machine, you may get the following error:
No space left on device. (refer here) - Solution:
export TMPDIR=$HOME/tmp(i.e. change /tmp directory location)mkdir -p $TMPDIRsource ~/.bashrc, and then run the following command -conda env create -f environment.yml
- While creating an env. on a linux machine, you may get the following error:
- While training:
- Some errors while training in a decentralized way:
ImportError: sys.meta_path is None, Python is likely shutting down- Solution - NOT YET RESOLVED!
- Some errors while training in a decentralized way:
- Notebooks:
- Data
transmission rate(i.e, sending large-sized tensors to the nodes) may be slow. (refer this)
- Data
- OpenMined Welcome Page, high level organization and projects
- OpenMined full stack, well explained
- Understanding PyGrid and the use of data-centric FL
- OpenMined RoadMap
- What is PyGrid demo
- Iterative, DVC: Data Version Control - Git for Data & Models (2020) DOI:10.5281/zenodo.012345.
- iterative.ai
- DVC Tutorials
Under Development: Please note that the project is in its early development stage and all the features have not been tested yet.
-
I would like to thank all my mentors for taking the time to mentor me and for their invaluable suggestions throughout. I truly appreciate their constant trust and encouragement!
-
Open Bioinformatics Foundation admins, helpdesk and the whole community
-
OpenMined Community, for putting together such a beautiful tech stack and for their constant help throughout!
-
Systems Biology of Aging Group, for providing me with useful resources, for trusting me throughout and for their constant feedback!
-
Iterative.ai and DVC, for making all of our lives so much more easier now :)
-
GSoC organizers, managers and Google.