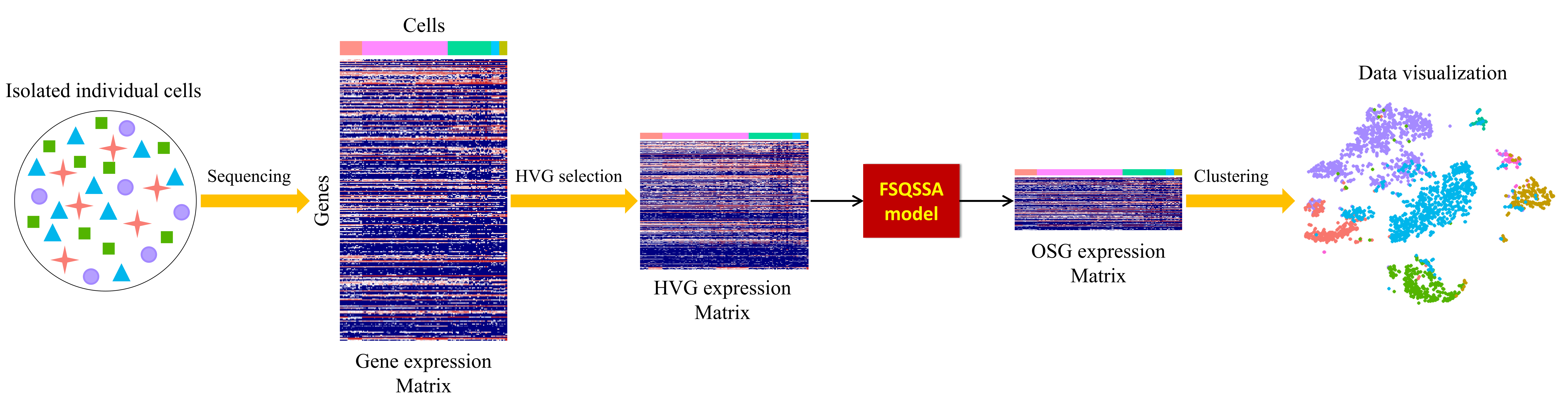
scFseCluster: a Feature selection enhanced clustering for single cell RNA-seq data.
With scFseCluster package, you can:
- Preprocess single cell gene expression.
- Obtain the optimal subset of features.
- Obtain clustering assignments of cells.
- Visualize cell clustering results.
For more about scFseCluster, please see details in our paper: scFseCluster: a Feature selection enhanced clustering for single cell RNA-seq data.
To use scFseCluster, you must make sure that your Python version is 3.7 or higher. If you don’t know the version of python you can check it by:
python
>>> import platform
>>> platform.python_version()
'3.7.11'We recommend that you install the environment that scFseCluster depends on in Anaconda (see Installing Anaconda). After installing Anaconda, you can create a new environment, for example, scFseCluster (you can change to any name you like):
# create an environment called scFseCluster
conda create -n scFseCluster python=3.7
# activate your enviorment
conda activate scFseCluster
# The install the dependencies needed for scFseCluster
pip install scikit_learn==0.22.1
pip install scanpy==1.4.6
# Because scFseCluster uses the R to draw diagrams, you need to install some packages for the R
conda install -c conda-forge r-base
conda install -c conda-forge r-seuratFinally, download scFseCluster from Github.
git clone https://github.com/wzqwtt/scFseClusterV1.0.git
cd scFseClusterIn the "scRNA-seq data" folder, we provide the Goolam dataset used in the paper as well as a description of each file in the dataset. For more datasets, please go to our website: http://cdsic.njau.edu.cn/data/scFseClusterV1.0.
scFseCluster is very easy to use. All you need to do is locate the scFseCluster directory and enter the following command to run it:
python scFseCluster.py --data dataset_pathdataset_path indicates the absolute path or relative path to specify a scRNA-seq expression matrix.
scFseCluster offers a range of scalable features. To use more parameters you can enter at the command line:
python scFseCluster.py --helpIf you want to use scFseCluster in jupyter notebook, you can check out the tutorials we provide.
Using the Goolam dataset as an example, run it using the command line as follows.
python scFseCluster.py --data ../scRNA-seq data/Goolam/raw_data.csv --plot TrueAfter entering this command scFseCluster will run. After running, three files will be output.
result.csv: This file stores the fitness values and the number of features generated in each iteration.subset.csv: This file stores the optimal subset of features after FSQSSA filtering.subset_index.csv: This file stores the sequence numbers of the filtered genes.
Please consider citing the following reference:
- scFseCluster: a Feature selection enhanced clustering for single cell RNA-seq data.