Welcome to the Vesalius GitHub page!
UNDER CONSTRUCTION - NEW VERSION COMMING SOON
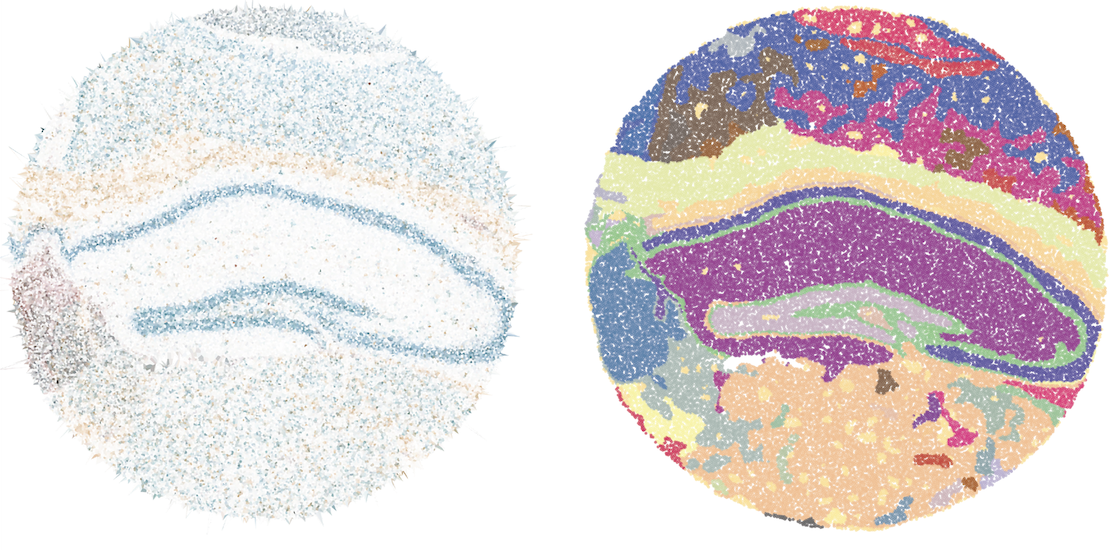
Vesalius is an R package to decipher tissue anatomy by embracing various image analysis techniques for high-resolution ST data. Vesalius identifies spatially expressed genes linked to the morphology of tissue structures.
If you do not have devtools already installed, please do so beforehand.
install.packages("devtools")
Ensure that the library has been loaded
library(devtools)
Install Vesalius via GitHub
install_github("WonLab-CS/Vesalius")
Vesalius provides an internal data set taken from real Spatial Transcriptomic data. This can be used as a dummy data set to get a feel for the Vesalius workflow.
The "Quick Start" guide can be found here
An in depth view of the Vesalius workflow can be found here. This contains the entire analysis related to the Vesalius Molecular Systems Biology.
NOTE: The in depth analysis file, contains path to files that should be changed accordingly.
The Vesalius package is in its early stage of development. We would ask you to share with us any bugs, concerns, or features you wish to see implemented.
Please open a GitHub issue or send an email to Patrick Martin (Patrick.Martin@cshs.org - pcnmartin@gmail.com)