Website: jasminezhoulab/cfSNV/
cfSNV is an ultra-sensitive and accurate somatic SNV caller designed for cfDNA sequencing. Taking advantage of modern statistical models and machine learning approaches, cfSNV provides hierarchical mutation profiling and multi-layer error suppression, including error suppression in read mates, site-level error filtration and read-level error filtration. cfSNV can be freely used for educational and research purposes by non-profit institutions and U.S. government agencies only under the UCLA Academic Software License. For information on the use for a commercial purpose or by a commercial or for-profit entity, please contact Prof. Xiangong Jasmine Zhou (https://zhoulab.dgsom.ucla.edu/).
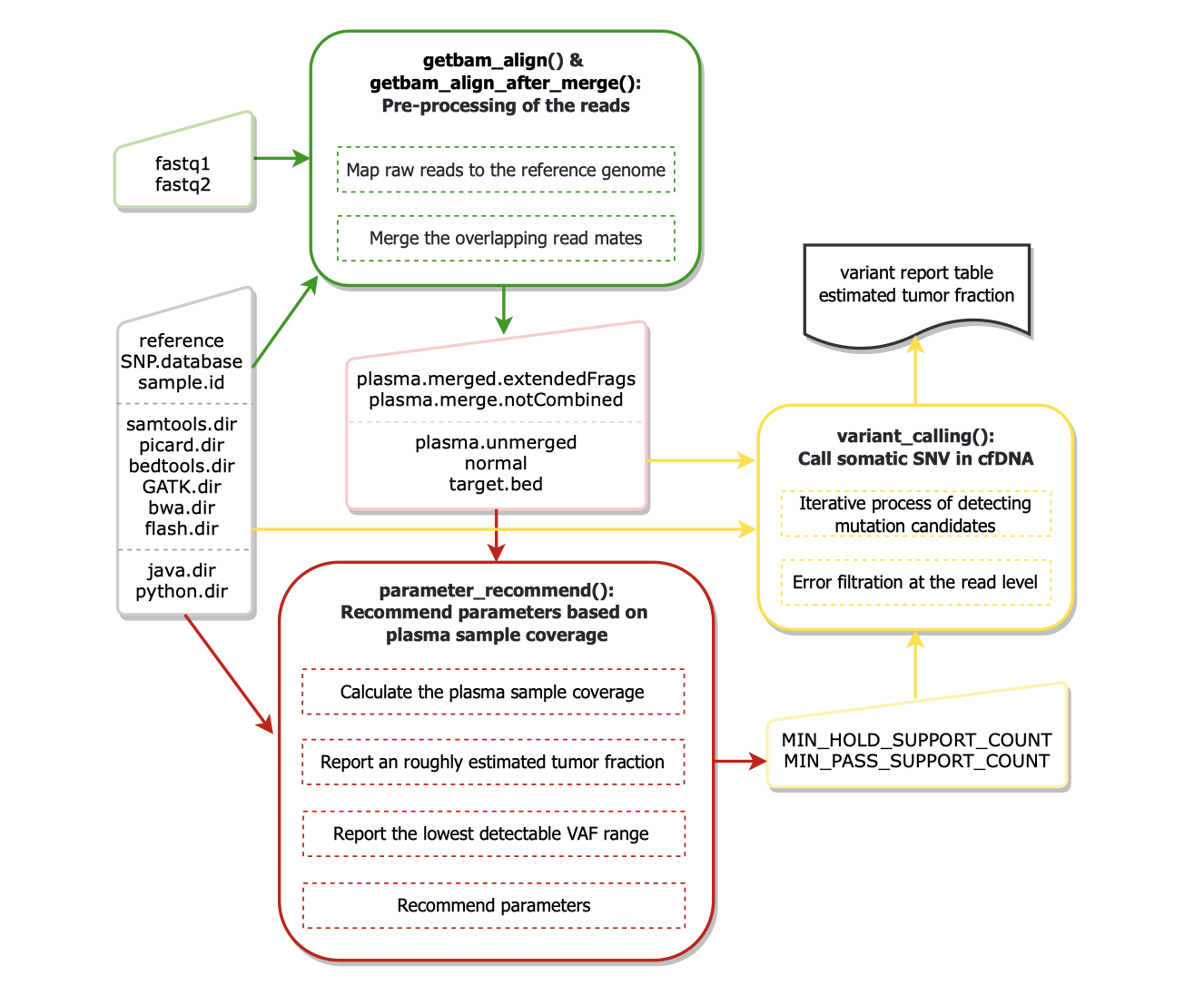
cfSNV provides a bioinformatics pipeline:
getbam_align()maps raw reads from fastq (gzip or not) files to the reference genome.getbam_align_after_merge()first merges the overlapping read mates in cfDNA sequencing data and then maps raw reads from fastq (gzip or not) files to the reference genome.parameter_recommend()recommends parameters based on plasma sample coverage.variant_calling()calls somatic SNV in cfDNA and reports estimated tumor fraction.
You can learn more about them in
vignette("cfSNV").
After downloading the cfSNV_0.99.0.tar.gz file to yourPath, you can
install cfSNV using the following
code:
install.packages("yourPath/cfSNV_0.99.0.tar.gz", repos = NULL, type = "source")- samtools 1.11: https://sourceforge.net/projects/samtools/files/samtools/
- picard 2.18.4: https://github.com/broadinstitute/picard/releases/tag/2.18.4
- bedtools 2.30.0: https://github.com/arq5x/bedtools2/releases/tag/v2.30.0
- GATK 3.8.0: https://github.com/broadgsa/gatk/releases/tag/3.8
- bwa 0.7.17: http://bio-bwa.sourceforge.net/
- FLASH2: https://github.com/dstreett/FLASH2
- java 1.8.0_281 (just need to be compatible with GATK and picard tools)
- python 3.7.0+
- numpy 1.13.3
- pandas 0.20.3
- scipy 1.1.0
- sklearn 0.24.1 (if using other versions, there will be a warining, but may not cause an error)
- R
4.0+
- Rcpp
- reticulate
- utils
library(cfSNV)
parameter_recommend(
plasma.unmerged, normal,
plasma.merged.extendedFrags, plasma.merge.notCombined,
target.bed, reference, SNP.database, samtools.dir, sample.id, roughly_estimated_tf = TRUE
)
#> The per base coverage of the plasma sample for each genomic region in the target bed file:
#> average = 105.222, median = 92.9562, 95th percentile = 227.395
#>
#> The roughly estimated tumor fraction in the plasma sample: 31.0295%
#> For a more accurate estimation, please run variant_calling().
#>
#> Lowest detectable VAF range under the default parameters: [2.199%, 5.277%]
#>
#> To detect different levels of lowest VAF,
#> at 1% VAF: MIN_HOLD_SUPPORT_COUNT = 8, MIN_PASS_SUPPORT_COUNT = 2;
#> at 5% VAF: MIN_HOLD_SUPPORT_COUNT = 17, MIN_PASS_SUPPORT_COUNT = 11
#> Note: decreasing the parameters (i.e. MIN_HOLD_SUPPORT_COUNT and MIN_PASS_SUPPORT_COUNT)
#> can lower the detection limit, but may also lower the variant quality.
results <- variant_calling(
plasma.unmerged, normal, plasma.merged.extendedFrags, plasma.merge.notCombined,
target.bed, reference, SNP.database, samtools.dir, picard.dir, bedtools.dir,
sample.id, MIN_HOLD_SUPPORT_COUNT, MIN_PASS_SUPPORT_COUNT
)
results$variant.list
#> CHROM POSITION ID REF ALT QUAL FILTER VAF
#> 1 chr22 20640684 . A G 2.918513e+08 PASS 0.3333333
#> 2 chr22 20640690 . G A 7.519190e+01 PASS 0.4666667
#> 3 chr22 20640765 . C G 1.000000e+00 PASS 0.0800000
#> 4 chr22 29075530 . T A 2.409049e+08 PASS 0.5882353
#> 5 chr22 29445285 . G A 3.205572e+28 PASS 0.2037037
#> 6 chr22 44342205 . T A 1.484423e+77 PASS 0.4285714
#> 7 chr22 44965238 . G A 3.889089e+23 PASS 0.1702128
#> 8 chr22 45601484 . G C 4.112031e+33 PASS 0.6666667
#> 9 chr22 45723968 . C T 1.198760e+11 PASS 0.6400000
results$tumor.fraction
#> [1] "31.6137566137%"Shuo Li, Zorawar S. Noor, Weihua Zeng, Mary L. Stackpole, Xiaohui Ni, Yonggang Zhou, Zuyang Yuan, Wing Hung Wong, Vatche G. Agopian, Steven M. Dubinett, Frank Alber, Wenyuan Li, Edward B. Garon, and Xianghong J. Zhou. Sensitive detection of tumor mutations from blood and its application to immunotherapy prognosis. Nature Communications. 2021 Jul 7;12(1):4172. doi: 10.1038/s41467-021-24457-2. PMID: 34234141; PMCID: PMC8263778.