The repository is an official implementation of SaProt: Protein Language Modeling with Structure-aware Vocabulary.
If you have any questions about the paper or the code, feel free to raise an issue!
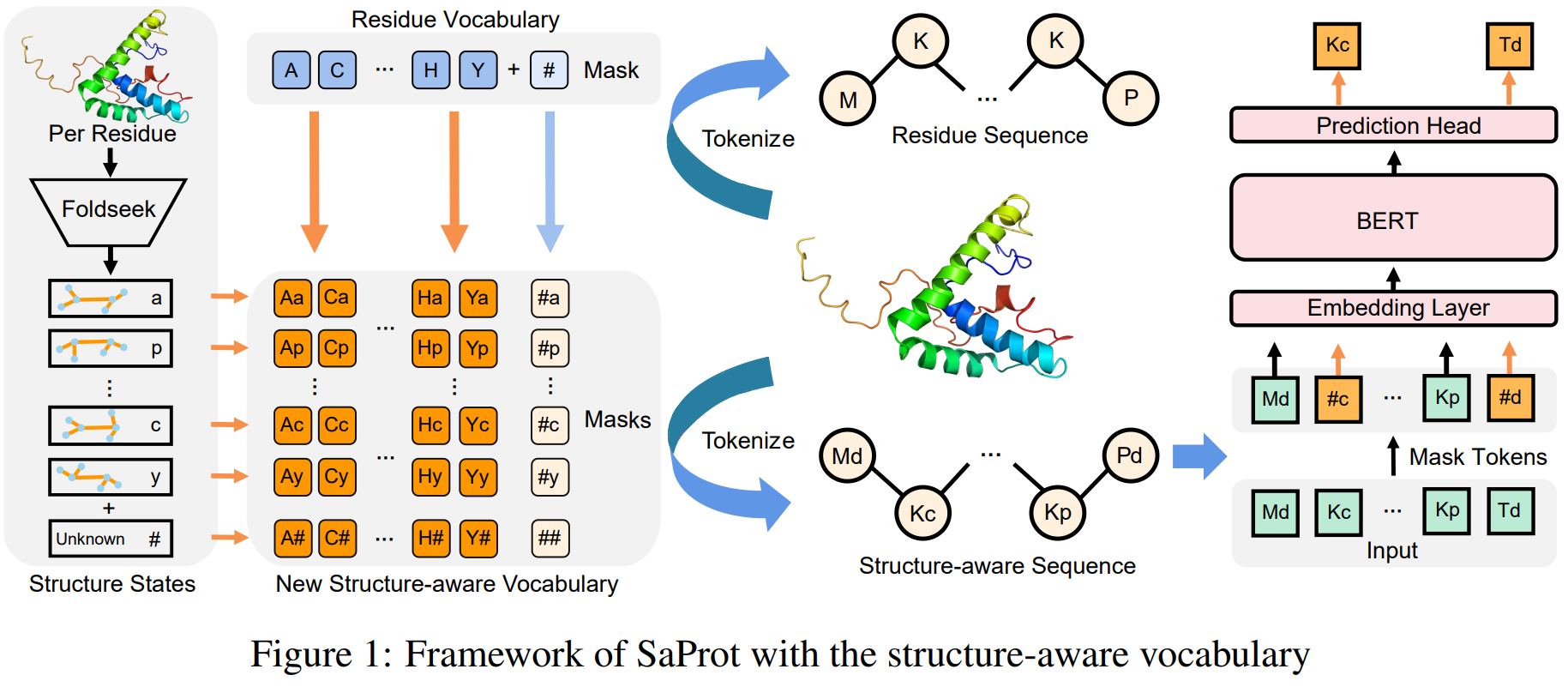
We propose a structure-aware vocabulary for protein language modeling. The vocabulary is constructed by encoding the
protein structure into discrete 3D tokens by using the foldseek. We combine the residue tokens and the structure tokens to form a structure-aware sequence.
Through large-scale pre-training, our model, i.e. SaProt, can learn the relationship between the structure and the sequence.
For more details, please refer to our paper https://www.biorxiv.org/content/10.1101/2023.10.01.560349v2.
conda create -n SaProt python=3.10
conda activate SaProt
bash environment.sh
We provide two ways to use SaProt, including through huggingface class and through the same way as in esm github. Users can choose either one to use.
We provide SaProt and SaProt-PDB for various use cases . The following code shows how to load the model.
from transformers import EsmTokenizer, EsmForMaskedLM
model_path = "/your/path/to/SaProt_650M_AF2"
tokenizer = EsmTokenizer.from_pretrained(model_path)
model = EsmForMaskedLM.from_pretrained(model_path)
#################### Example ####################
device = "cuda"
model.to(device)
seq = "MdEvVpQpLrVyQdYaKv"
tokens = tokenizer.tokenize(seq)
print(tokens)
inputs = tokenizer(seq, return_tensors="pt")
inputs = {k: v.to(device) for k, v in inputs.items()}
outputs = model(**inputs)
print(outputs.logits.shape)
"""
['Md', 'Ev', 'Vp', 'Qp', 'Lr', 'Vy', 'Qd', 'Ya', 'Kv']
torch.Size([1, 11, 446])
"""
The esm version is also stored in the same huggingface folder, named SaProt_650M_AF2.pt. We provide a function to load the model.
from utils.esm_loader import load_esm_saprot
model_path = "/your/path/to/SaProt_650M_AF2.pt"
model, alphabet = load_esm_saprot(model_path)
We provide a function to convert a protein structure into a structure-aware sequence. The function calls the
foldseek
binary file to encode the structure. You can download the binary file from here and place it in the bin folder
. The following code shows how to use it.
from utils.foldseek_util import get_struc_seq
pdb_path = "example/8ac8.cif"
# Extract the "A" chain from the pdb file and encode it into a struc_seq
# pLDDT is used to mask low-confidence regions if "plddt_path" is provided
parsed_seqs = get_struc_seq("bin/foldseek", pdb_path, ["A"])["A"]
seq, foldseek_seq, combined_seq = parsed_seqs
print(f"seq: {seq}")
print(f"foldseek_seq: {foldseek_seq}")
print(f"combined_seq: {combined_seq}")
We are preparing the pre-training dataset and will release it once our paper is accepted.
We provide datasets that are used in the paper. Datasets can be downloaded from here.
Once downloaded, the datasets need to be decompressed and placed in the LMDB folder for supervised fine-tuning.
We provide a script to fine-tune SaProt on the datasets. The following code shows how to fine-tune SaProt on specific
downstream tasks. Before running the code, please make sure that the datasets are placed in the LMDB folder and the
huggingface version of SaProt model is placed in the weights/PLMs folder. Note that the default training setting is not as
same as in the paper because of the hardware limitation for different users. We recommend users to modify the yaml file
flexibly based on their own conditions (i.e. batch_size, devices and accumulate_grad_batches).
# Fine-tune SaProt on the Thermostability task
python scripts/training.py -c config/Thermostability/saprot.yaml
# Fine-tune ESM-2 on the Thermostability task
python scripts/training.py -c config/Thermostability/esm2.yaml
If you want to record the training process using wandb, you could modify the config file and set Trainer.logger = True
and then paste your wandb API key in the config key setting.os_environ.WANDB_API_KEY.
We provide a script to evaluate the zero-shot performance of models (foldseek binary file is required to be placed in
the bin folder):
# Evaluate the zero-shot performance of SaProt on the ProteinGym benchmark
python scripts/mutation_zeroshot.py -c config/ProteinGym/saprot.yaml
# Evaluate the zero-shot performance of ESM-2 on the ProteinGym benchmark
python scripts/mutation_zeroshot.py -c config/ProteinGym/esm2.yaml
The results will be saved in the output/ProteinGym folder.
For ClinVar benchmark, you can use the following script to calculate the AUC metric:
# Evaluate the zero-shot performance of SaProt on the ClinVar benchmark
python scripts/mutation_zeroshot.py -c config/ClinVar/saprot.yaml
python scripts/compute_clinvar_auc.py -c config/ClinVar/saprot.yaml
If you find this repository useful, please cite our paper:
@article{su2023saprot,
title={SaProt: Protein Language Modeling with Structure-aware Vocabulary},
author={Su, Jin and Han, Chenchen and Zhou, Yuyang and Shan, Junjie and Zhou, Xibin and Yuan, Fajie},
journal={bioRxiv},
year={2023},
publisher={Cold Spring Harbor Laboratory}
}