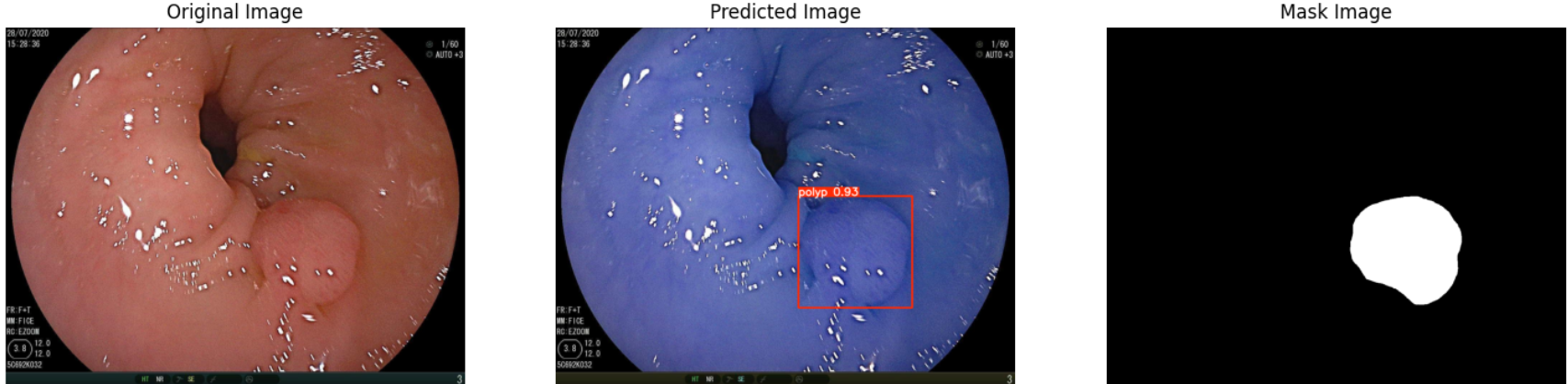
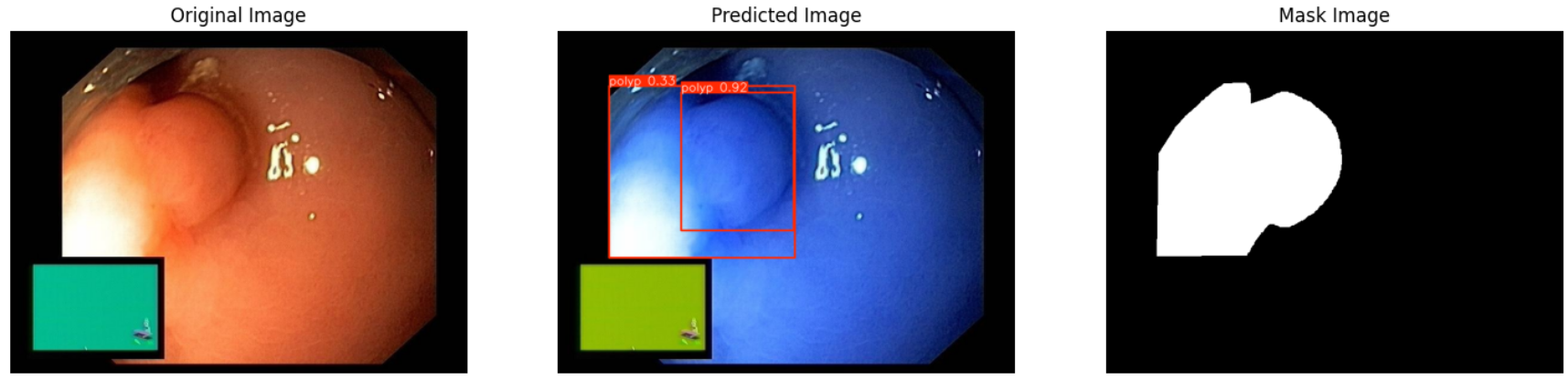
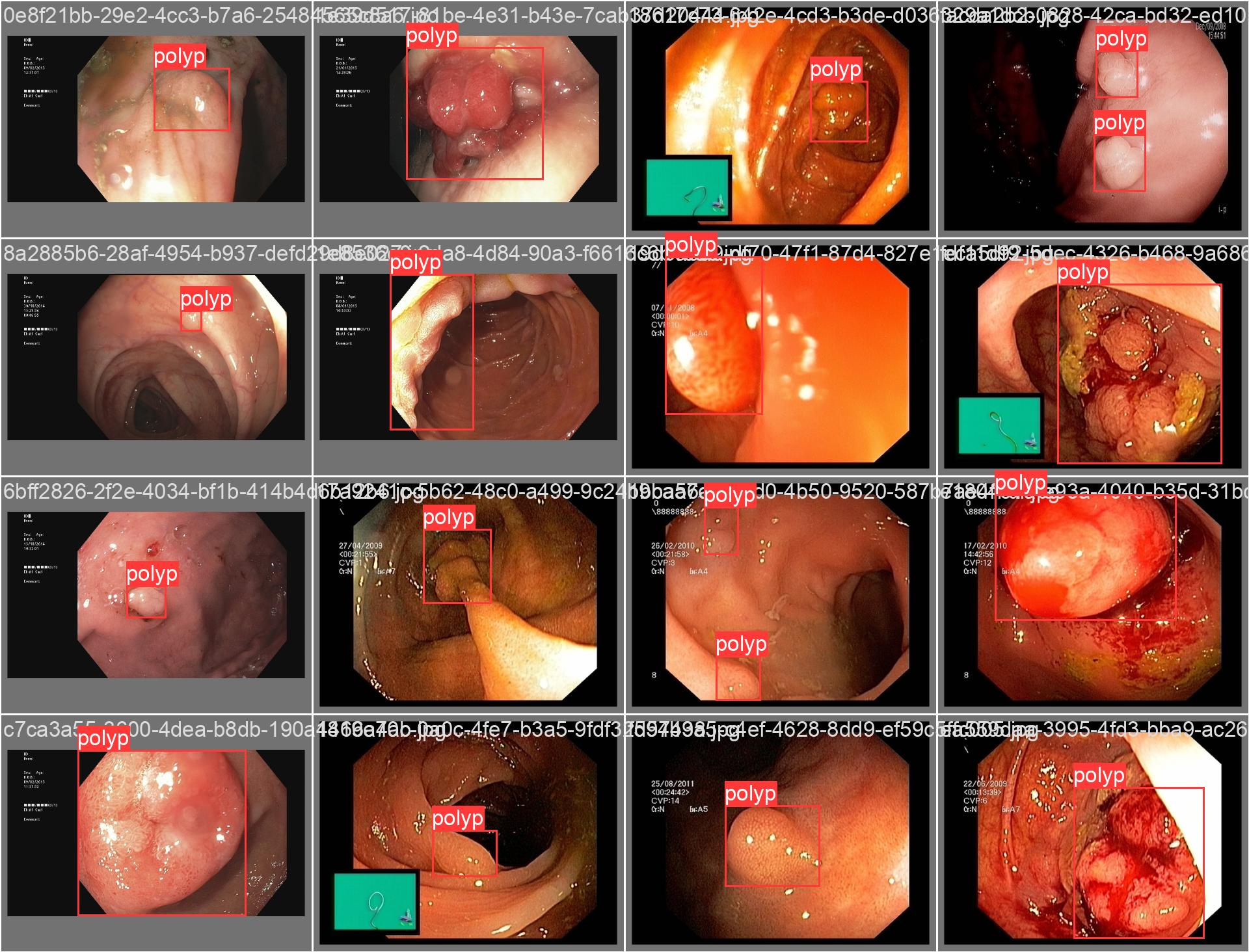
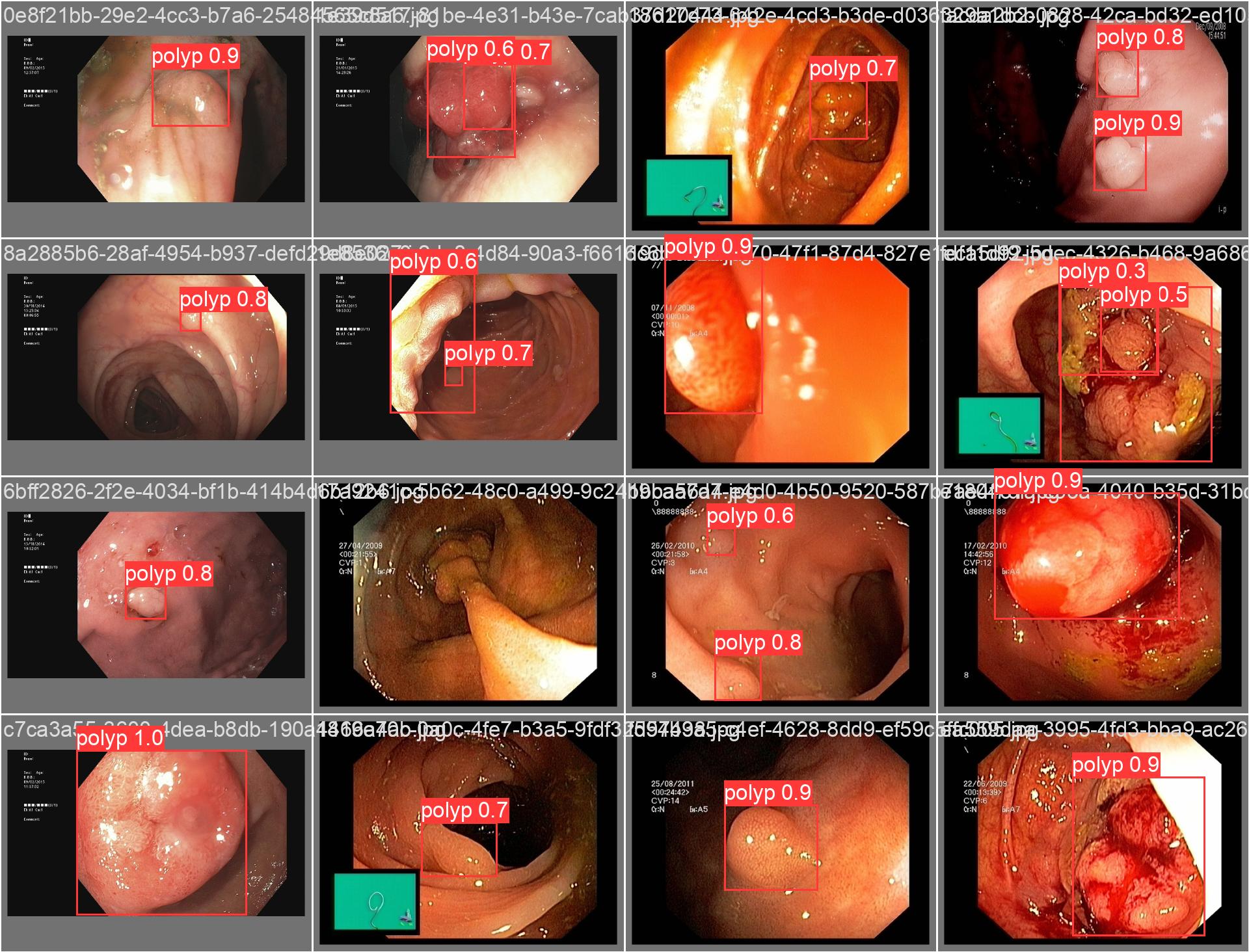
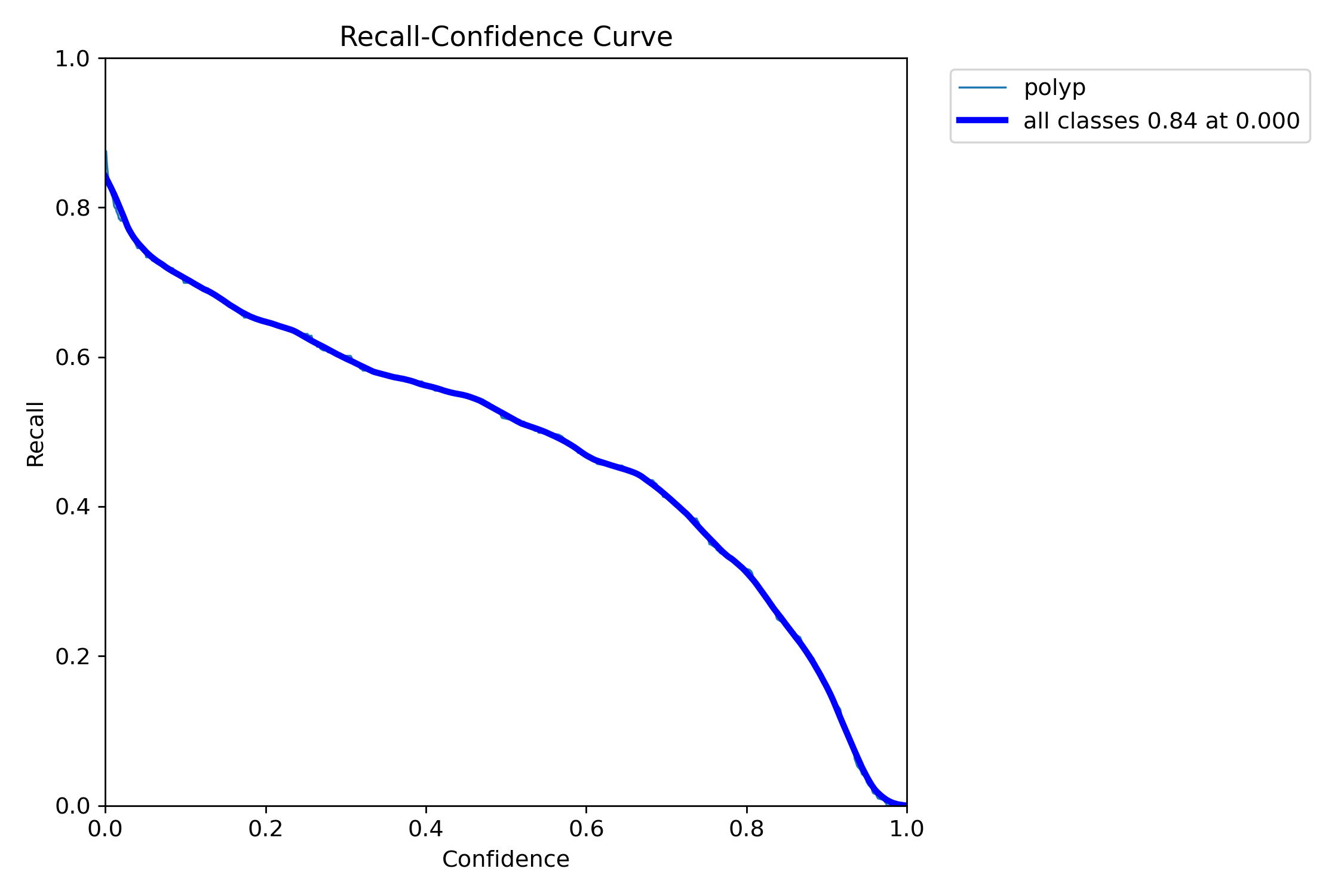
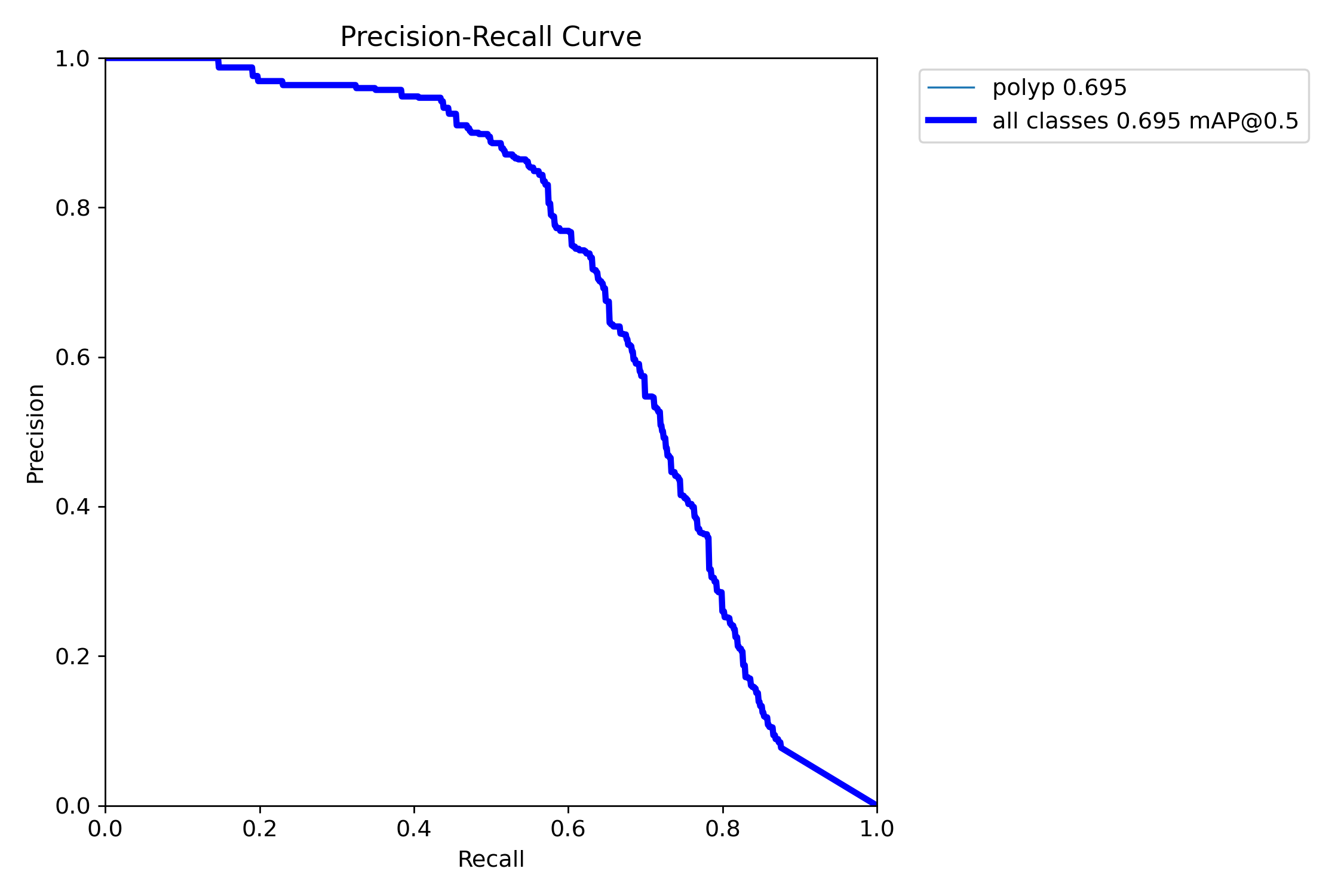
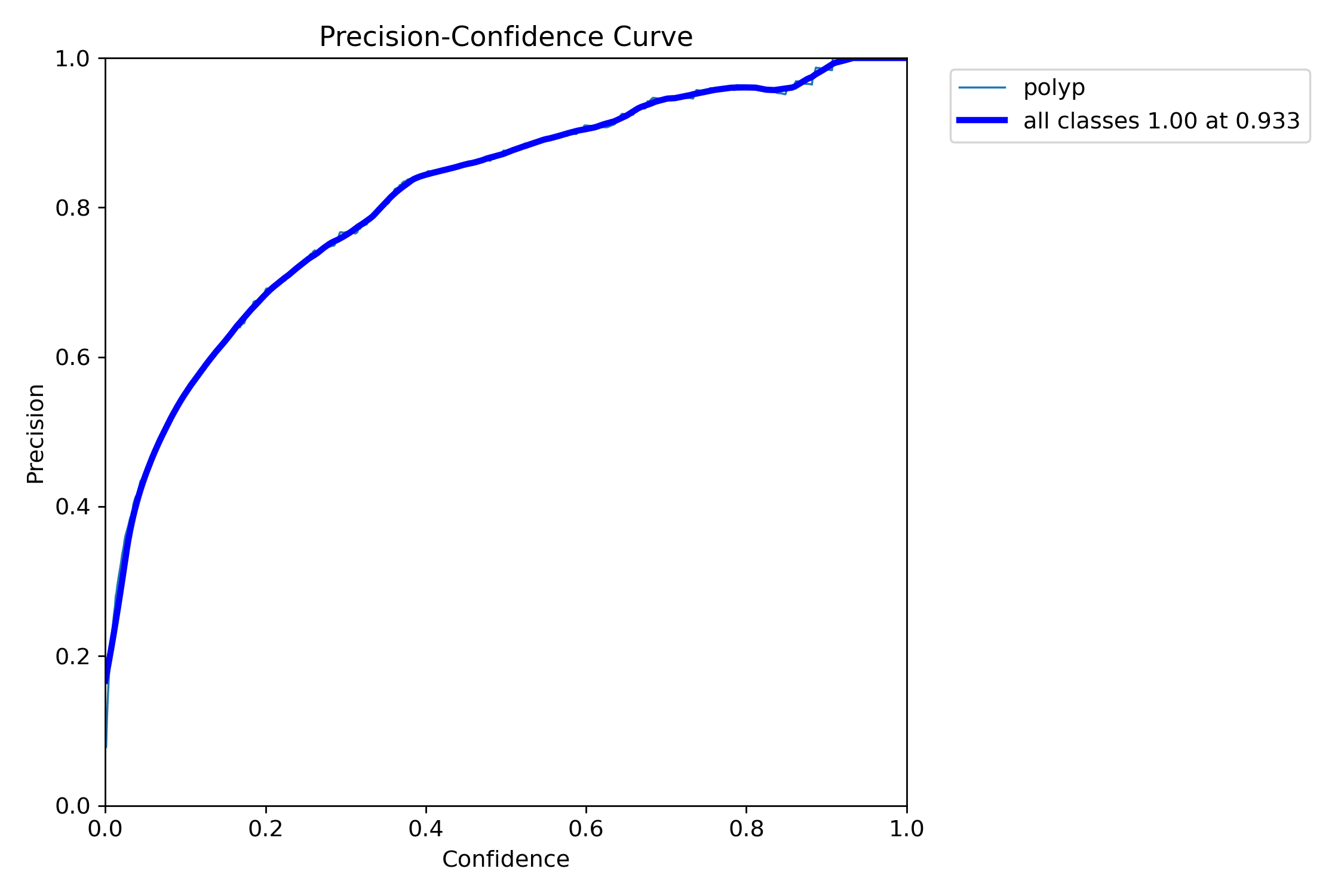
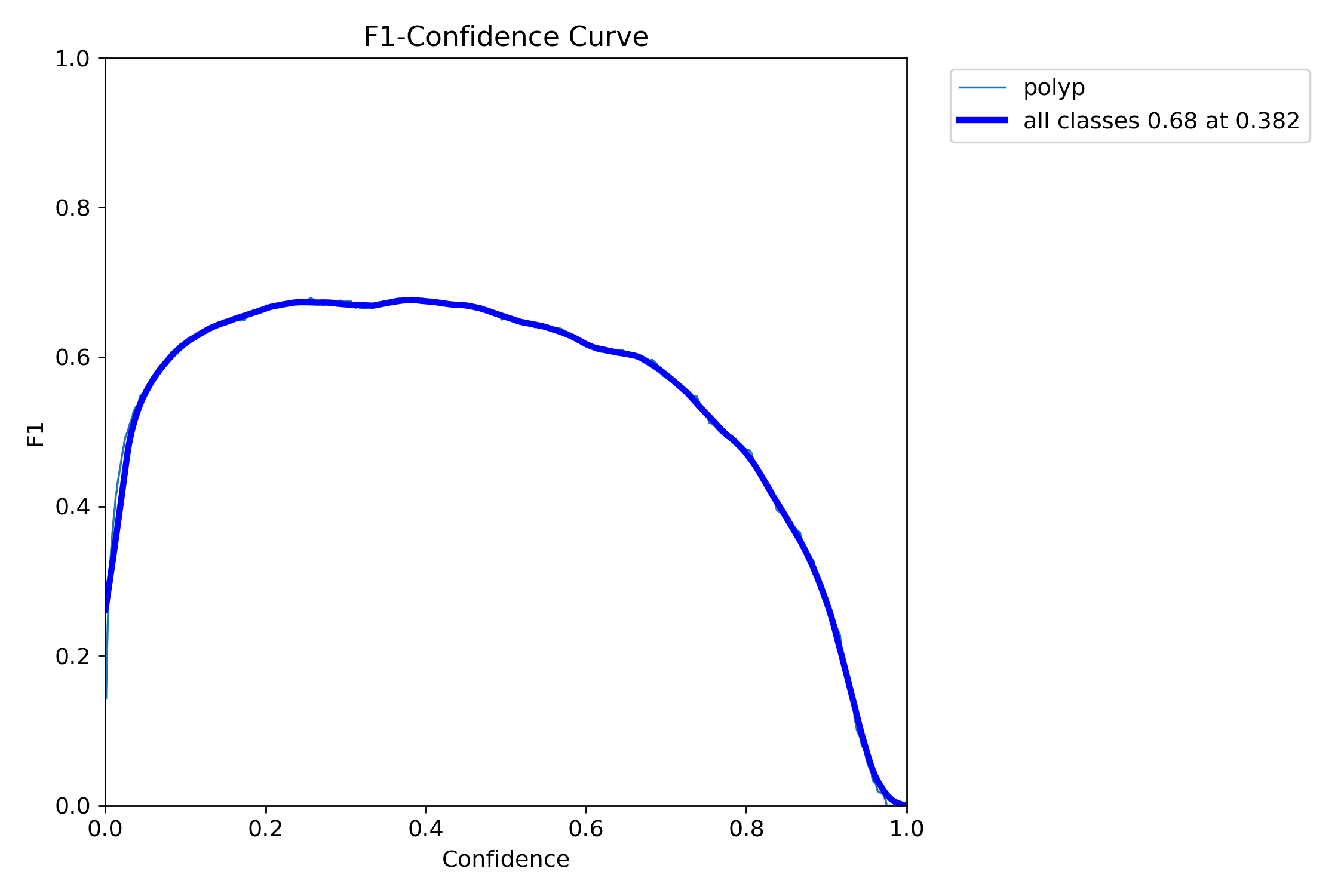
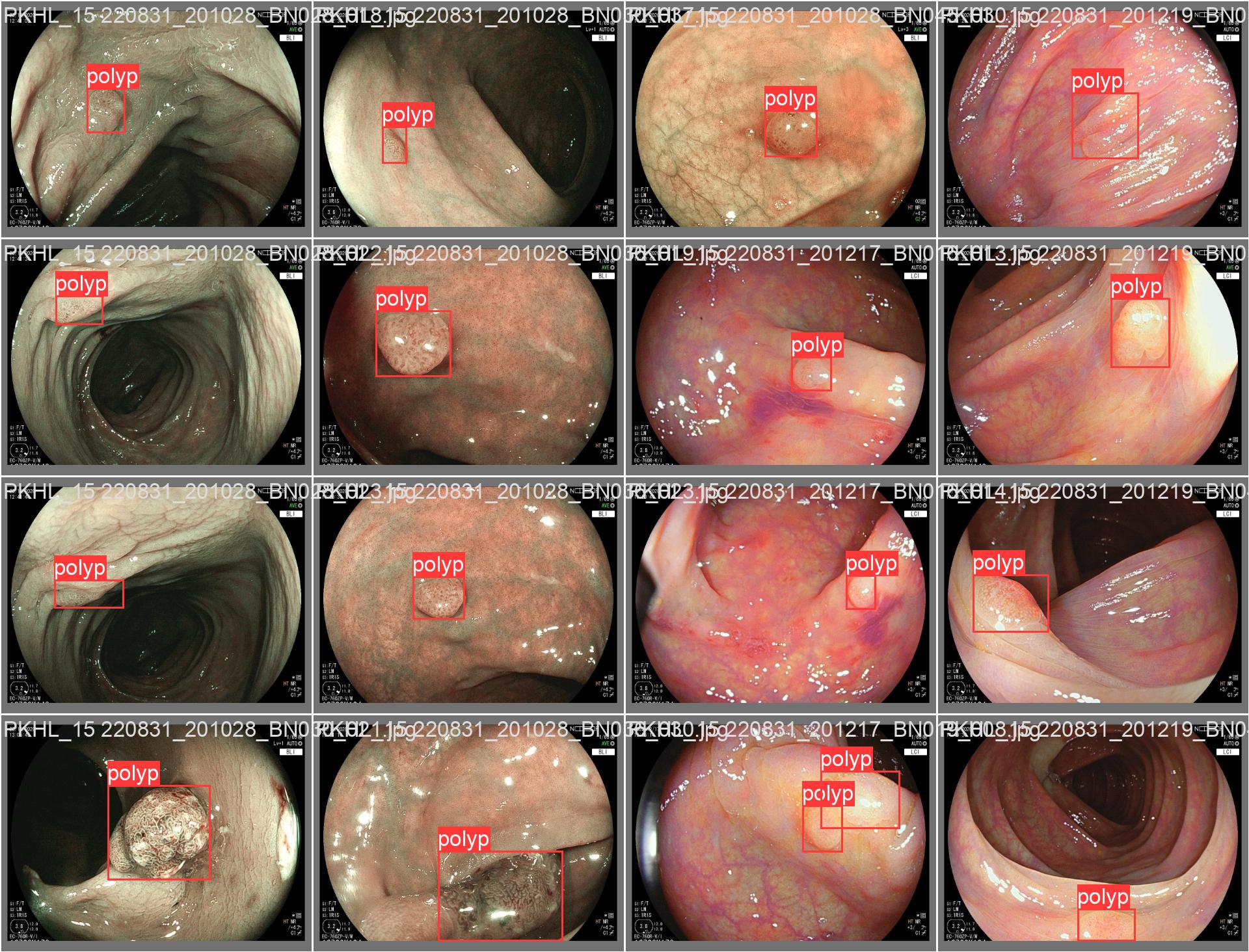
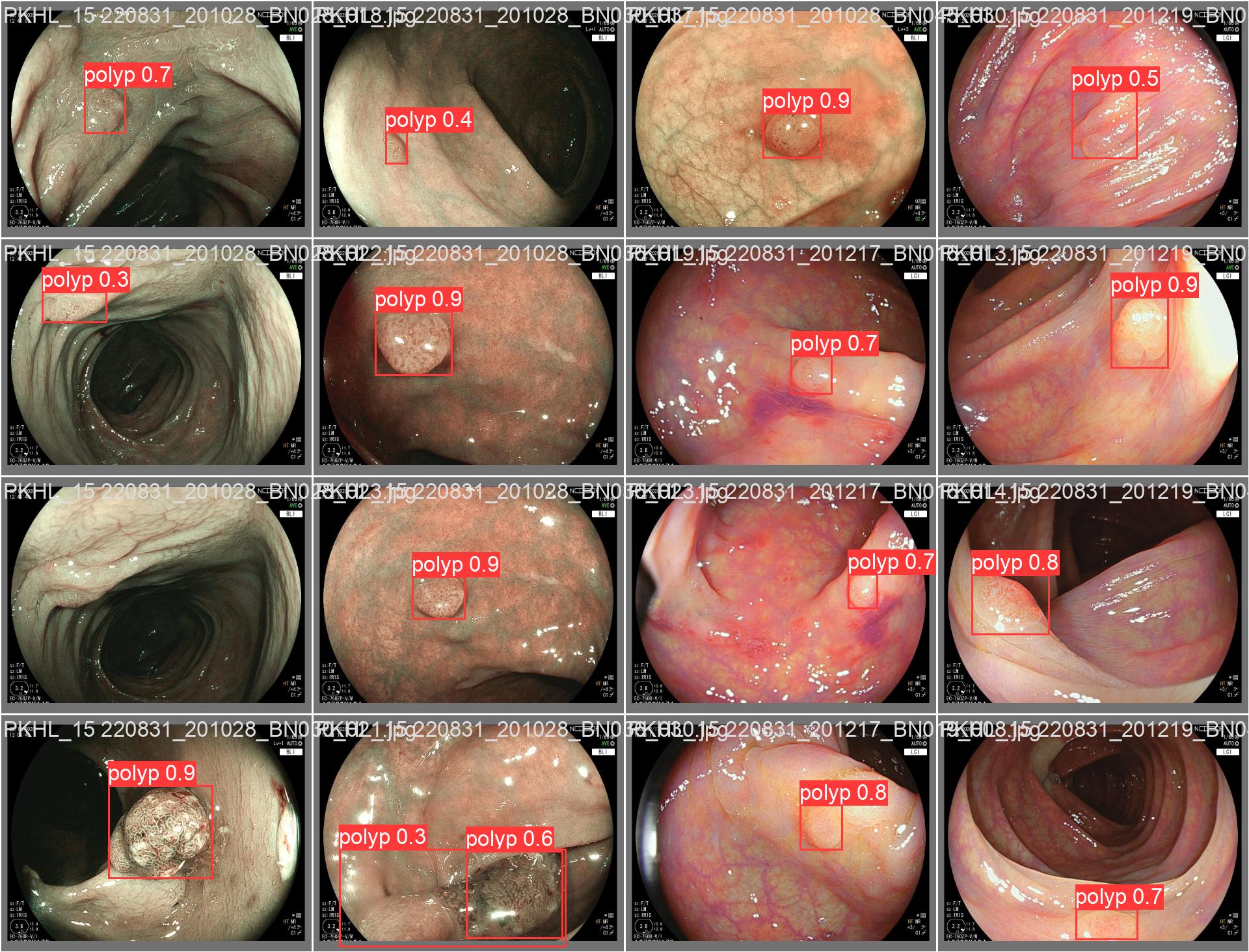
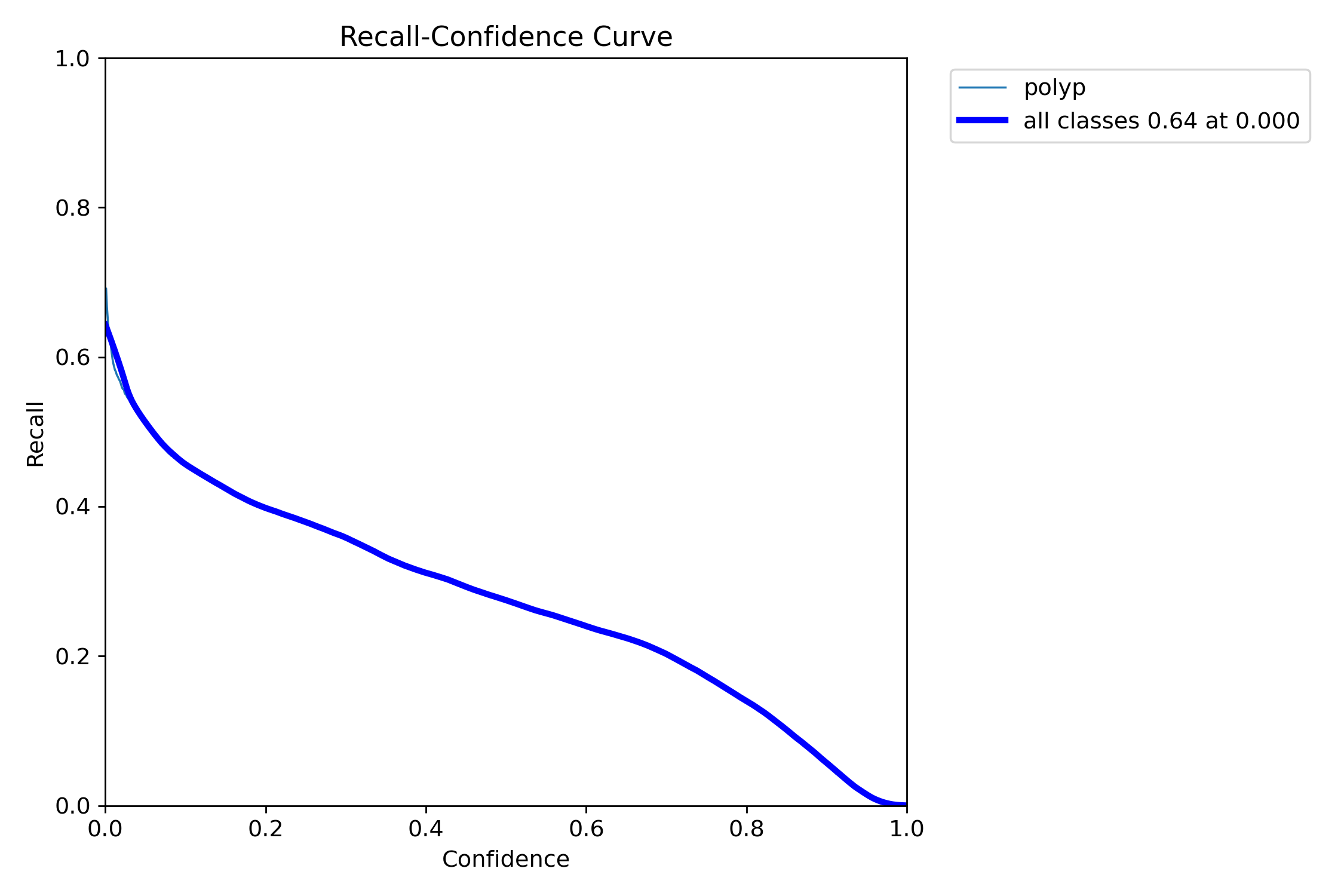
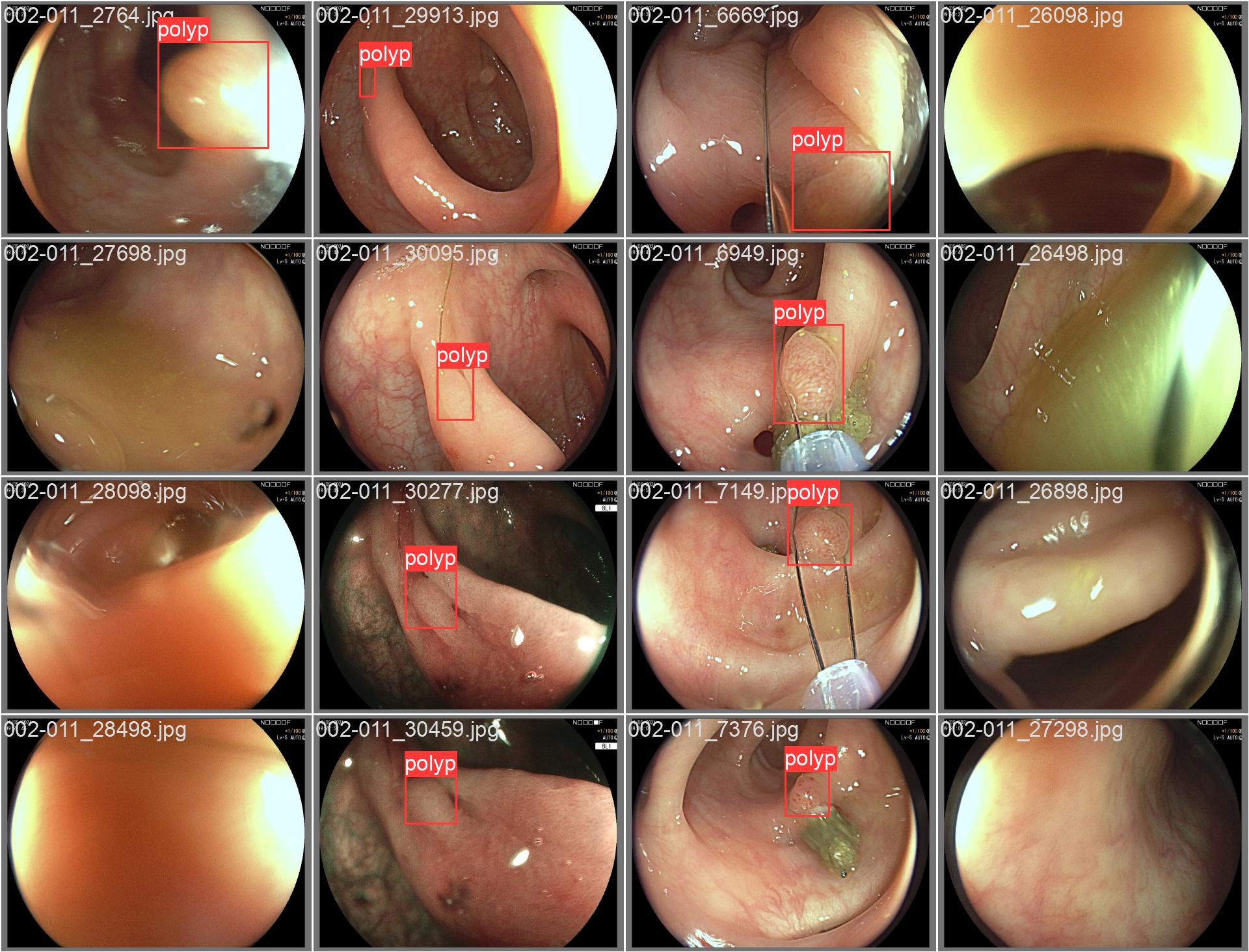
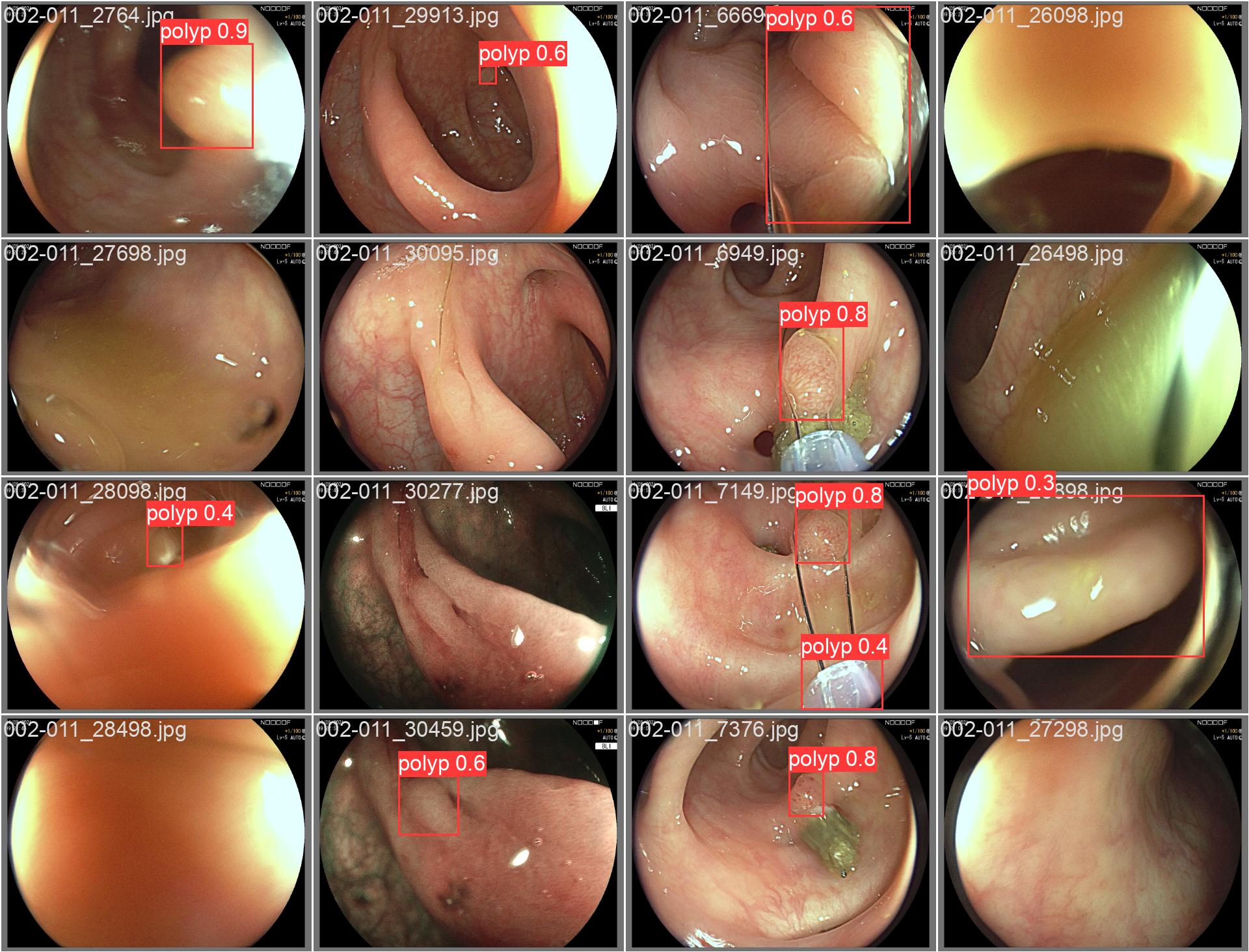
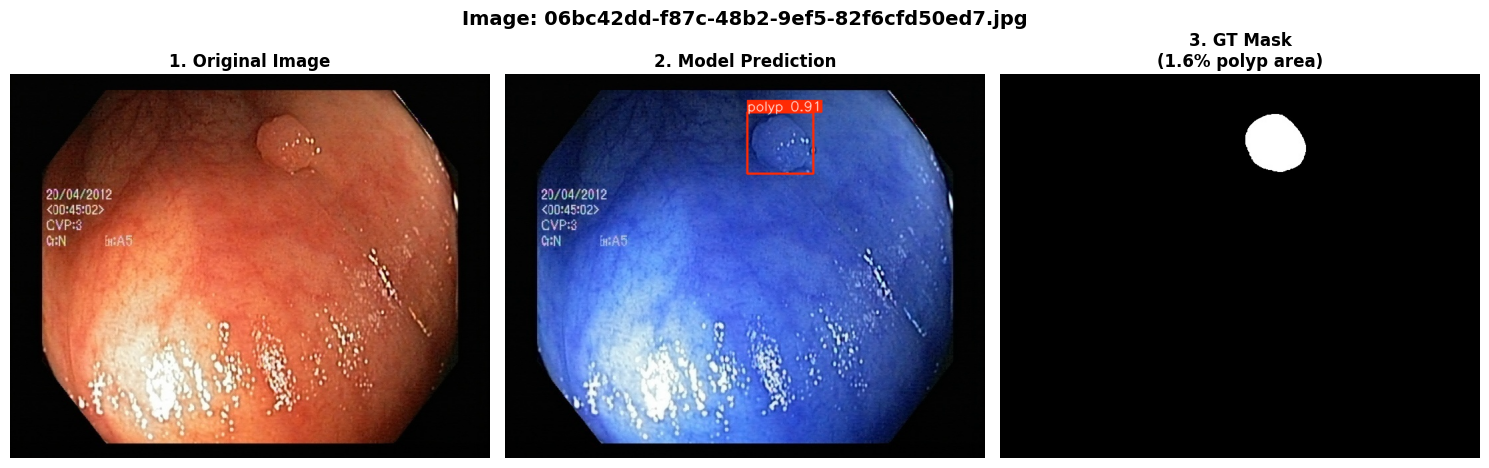
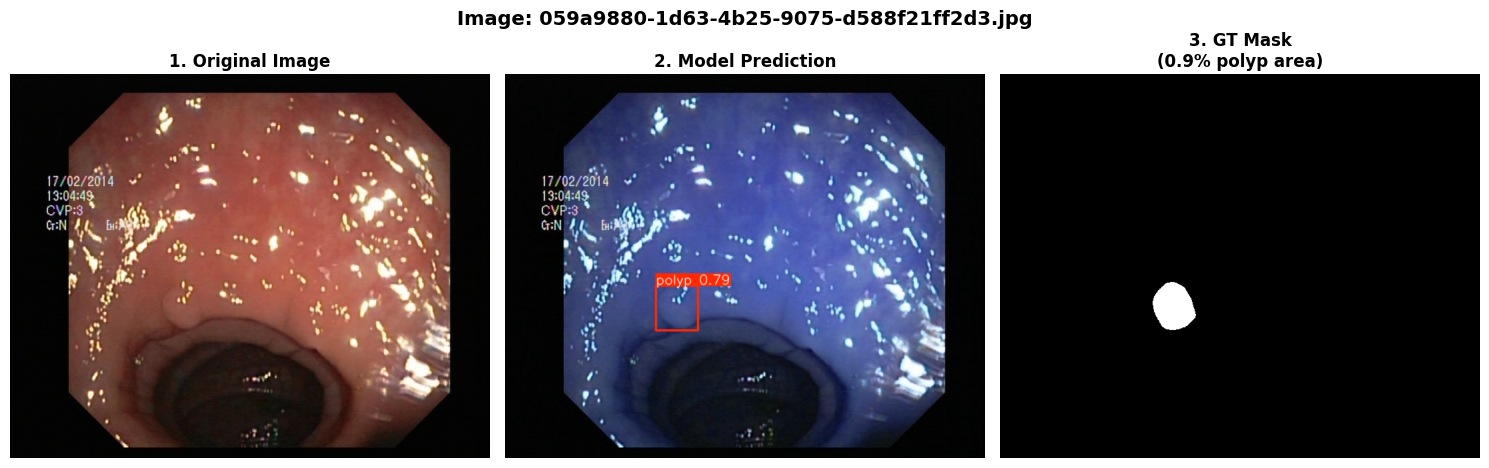
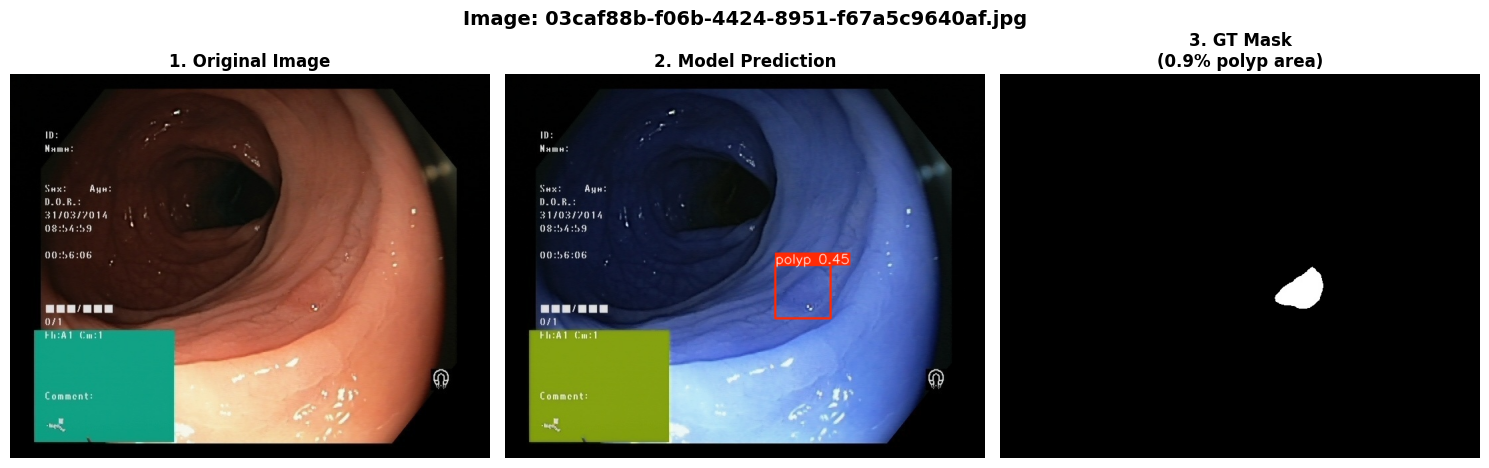
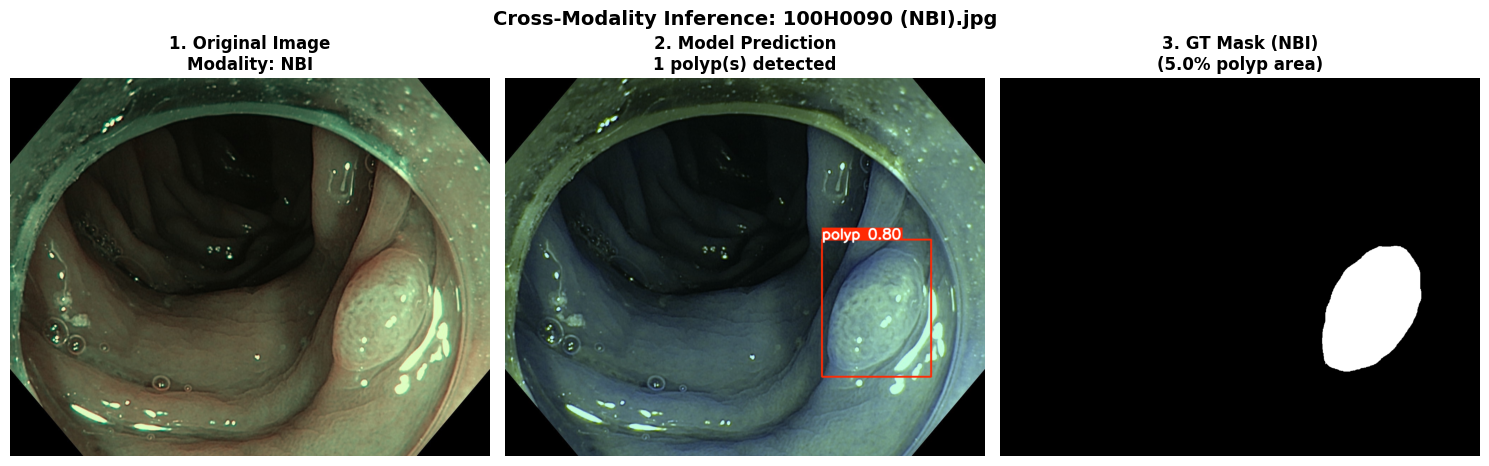
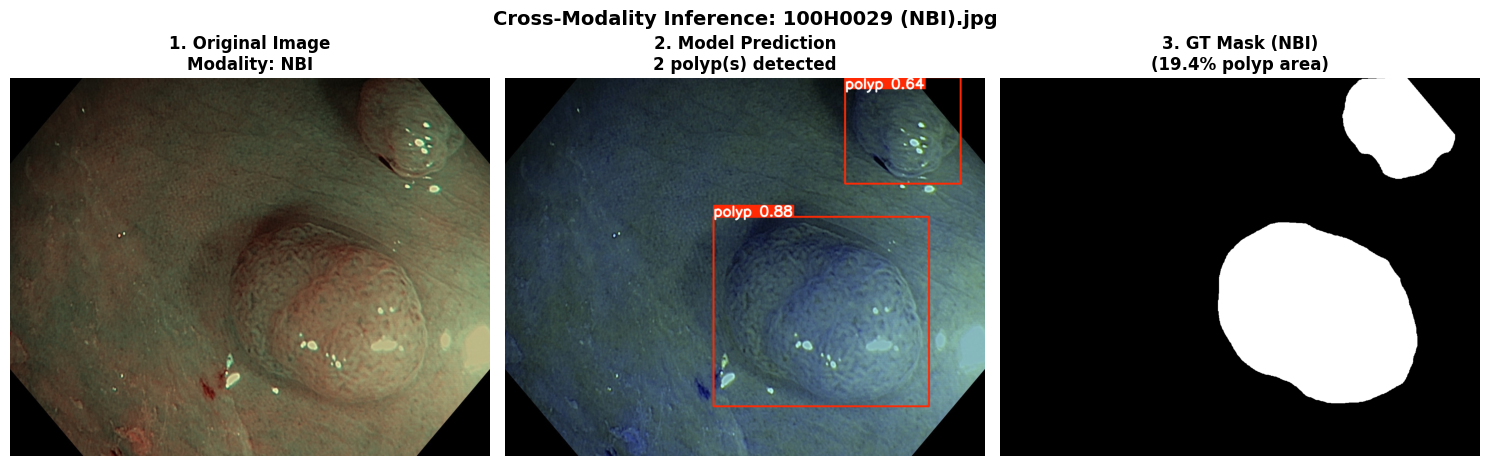
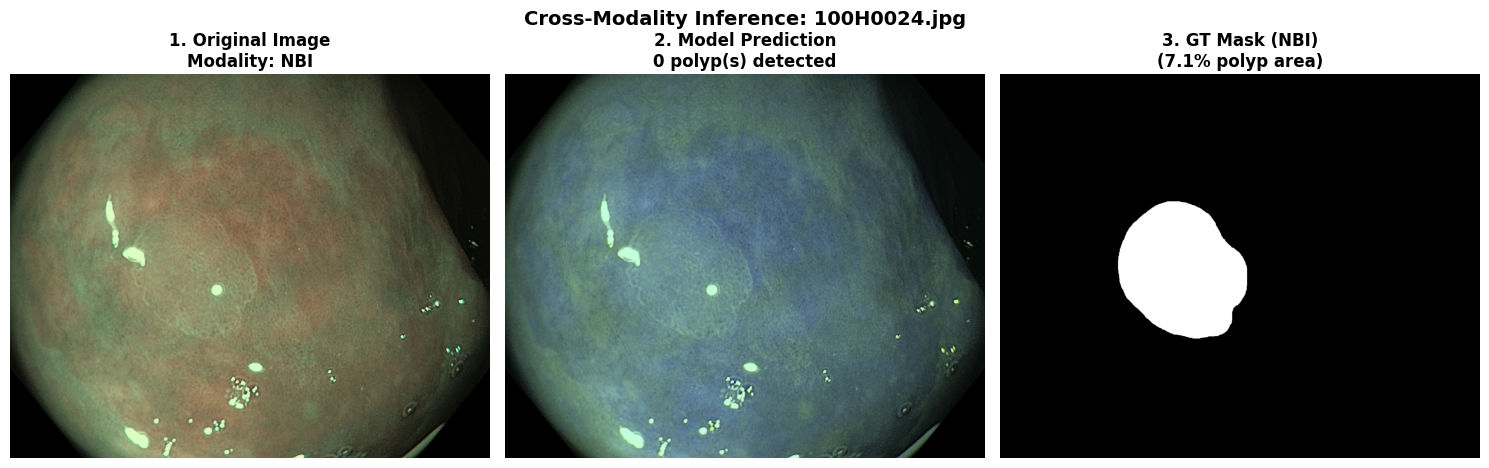
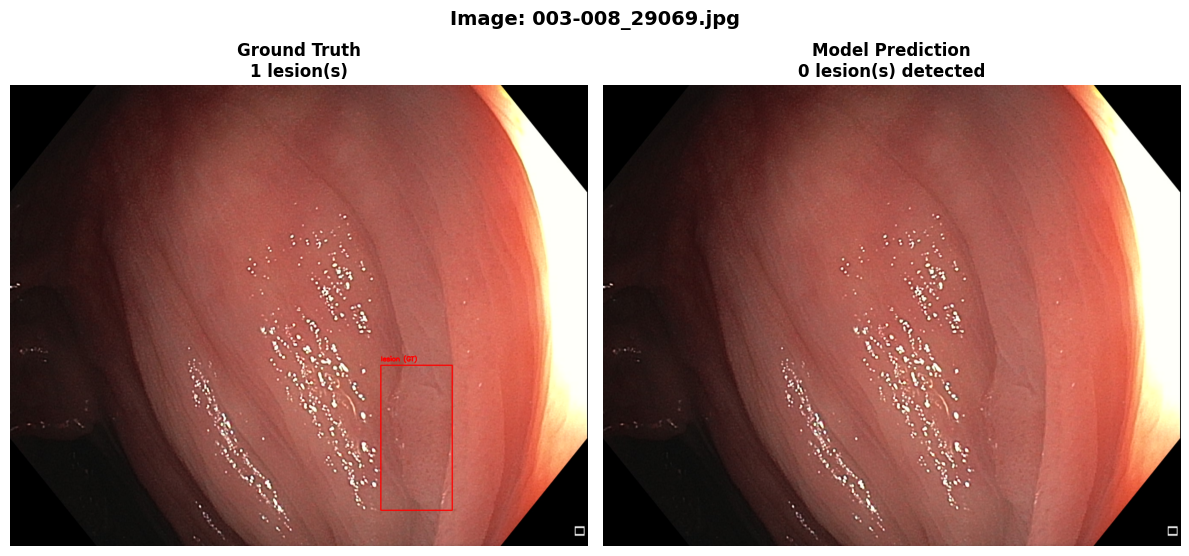
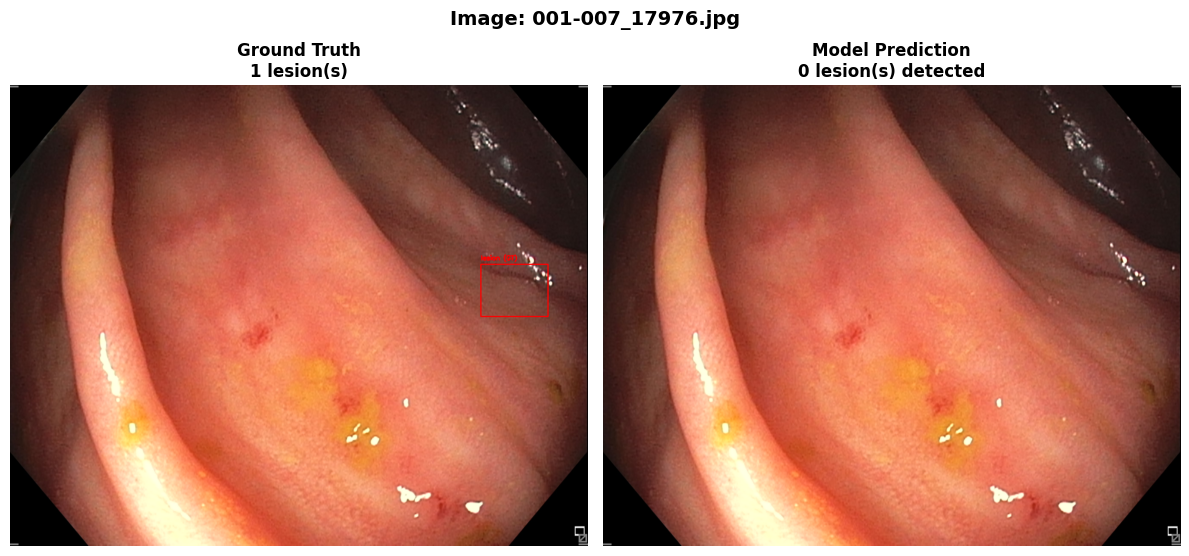
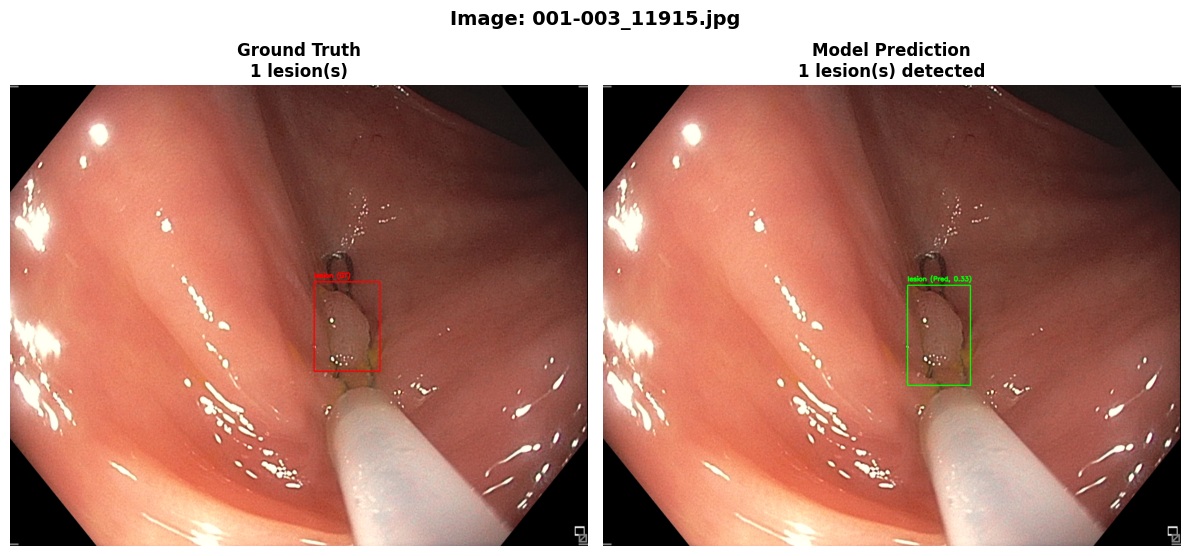
In this repo, we are going to employ Machine learning methods to detect colon tissues(polyp).
- WLI (White Light Imaging): Standard endoscopic view
- NBI (Narrow Band Imaging): Enhanced vascular pattern visualization
- LCI (Linked Color Imaging): Improved color contrast
- FICE (Flexible Spectral Imaging Color Enhancement): Spectral enhancement
- BLI (Blue Laser Imaging): Surface structure enhancement
REAL-Colon provides 60 full-resolution, real-world colonoscopy videos (2.7M frames) from multiple centers, with 350k expert-annotated polyp bounding boxes. Includes clinical metadata, acquisition details, and histopathology. Designed for robust CADe/CADx development and benchmarking. Released for non-commercial research. See the paper for details.
- Train
YOLOv9m summary (fused): 151 layers, 20,013,715 parameters, 0 gradients, 76.5 GFLOPs
Class Images Instances Box(P R mAP50 mAP50-95)
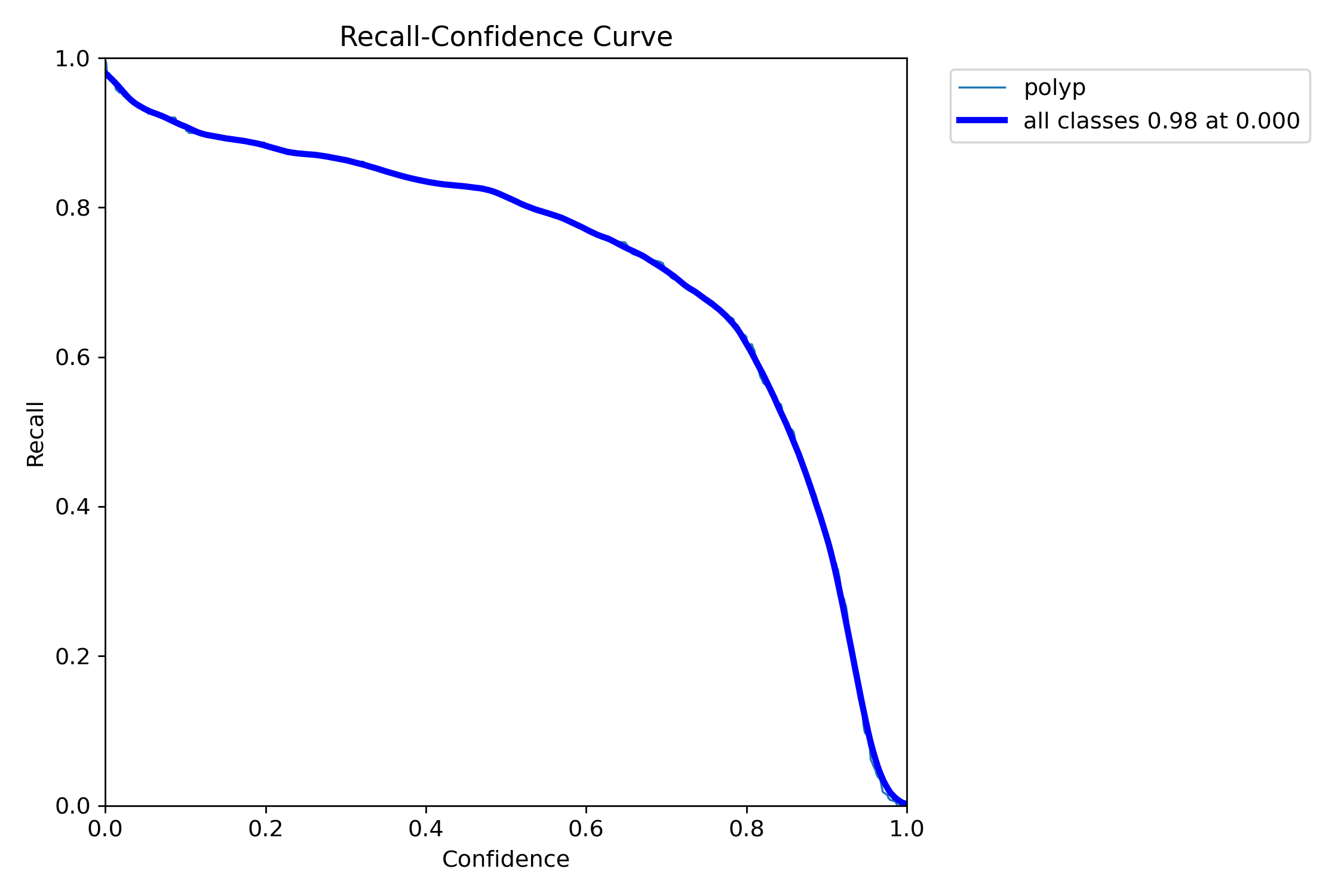
all 359 382 0.933 0.912 0.963 0.816- detect polyps(bounding box)
Mamba-YOLO merges the state-space modeling efficiency of Mamba with the real-time detection strength of YOLOv8.
The architecture replaces the CSP backbone with a Selective Scan (Mamba) block, enabling long-range spatial dependency modeling at reduced computational cost.
This implementation targets medical image analysis, specifically polyp detection from multimodal colonoscopy datasets.
| Component | Description |
|---|---|
| Backbone | Mamba-based state-space selective scan layers replacing CSP blocks |
| Neck | PANet-style feature pyramid |
| Head | YOLOv8 detection head (multi-scale anchors) |
| Losses | CIoU + BCE + objectness loss |
| Training Framework | Ultralytics YOLO API |
| Hardware | NVIDIA T4 (16 GB) × 2 |
| Software Stack | PyTorch 2.3.1 + CUDA 12.1, Python 3.11 |
| Parameter | Value |
|---|---|
| Modality | WLI |
| Epochs | 300 |
| Batch size | 16 |
| Optimizer | AdamW |
| Image size | 640×640 |
| Scheduler | Cosine annealing |
| Mixed precision | AMP enabled |
=== Test Set Metrics on trained modality(WLI)===
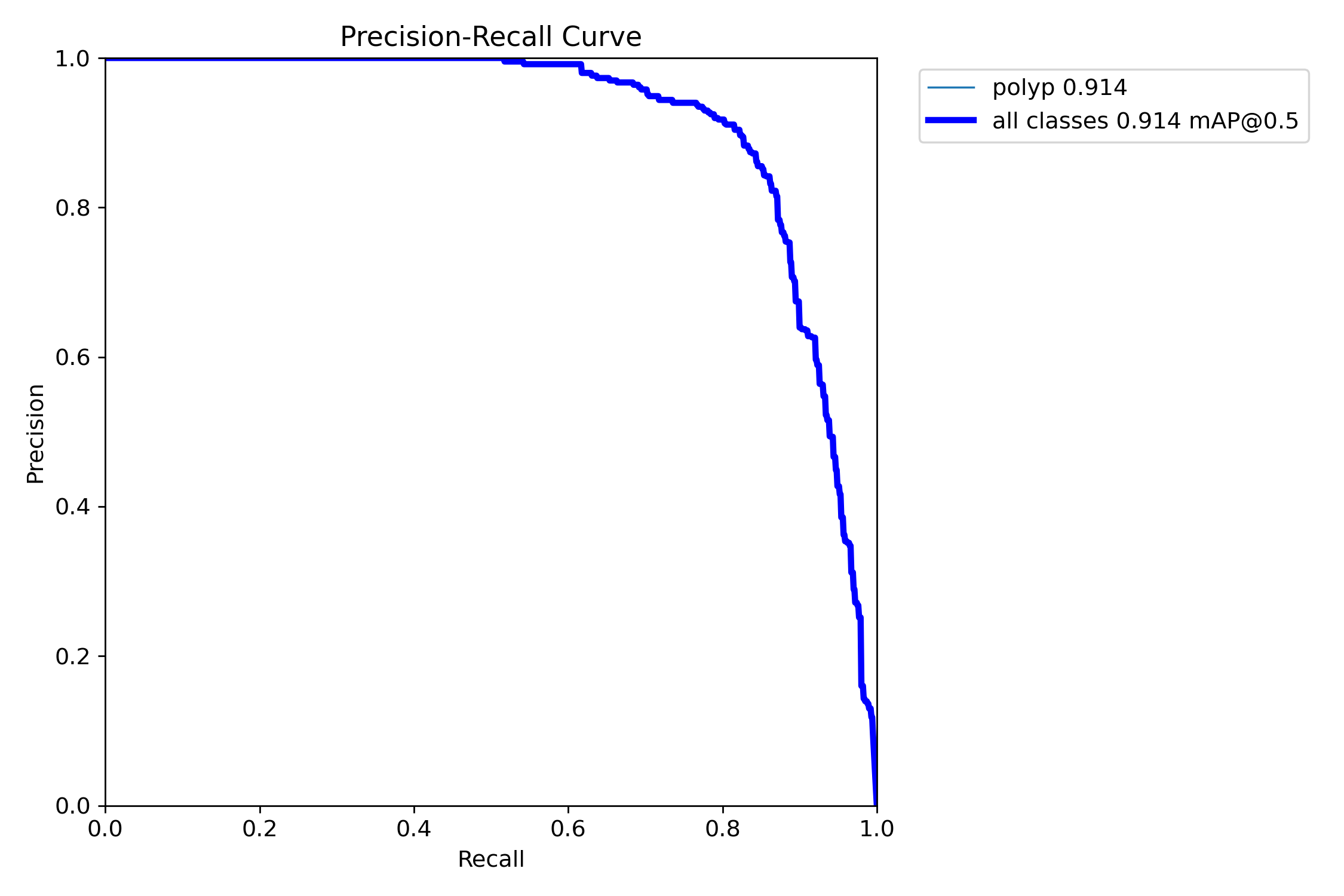
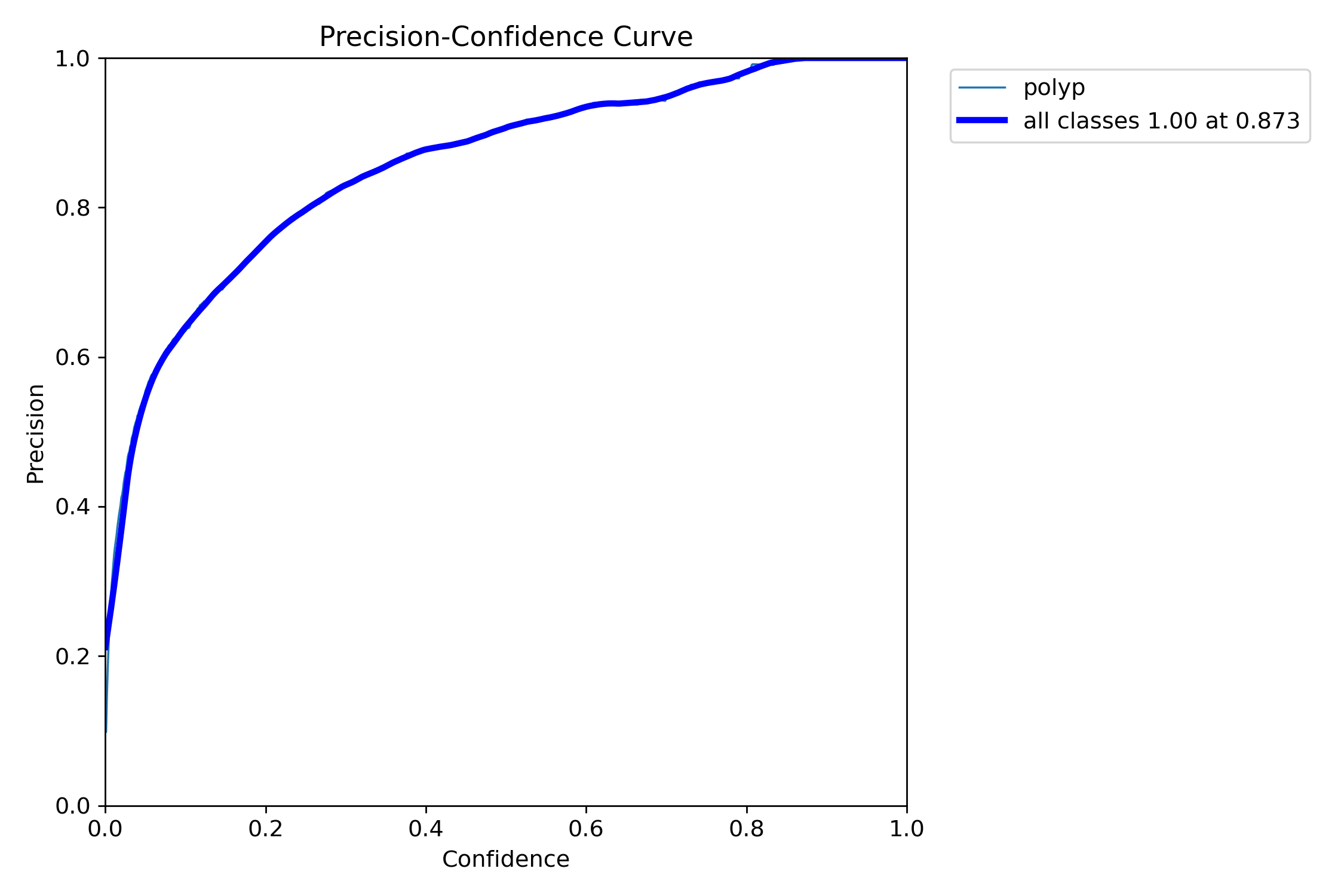
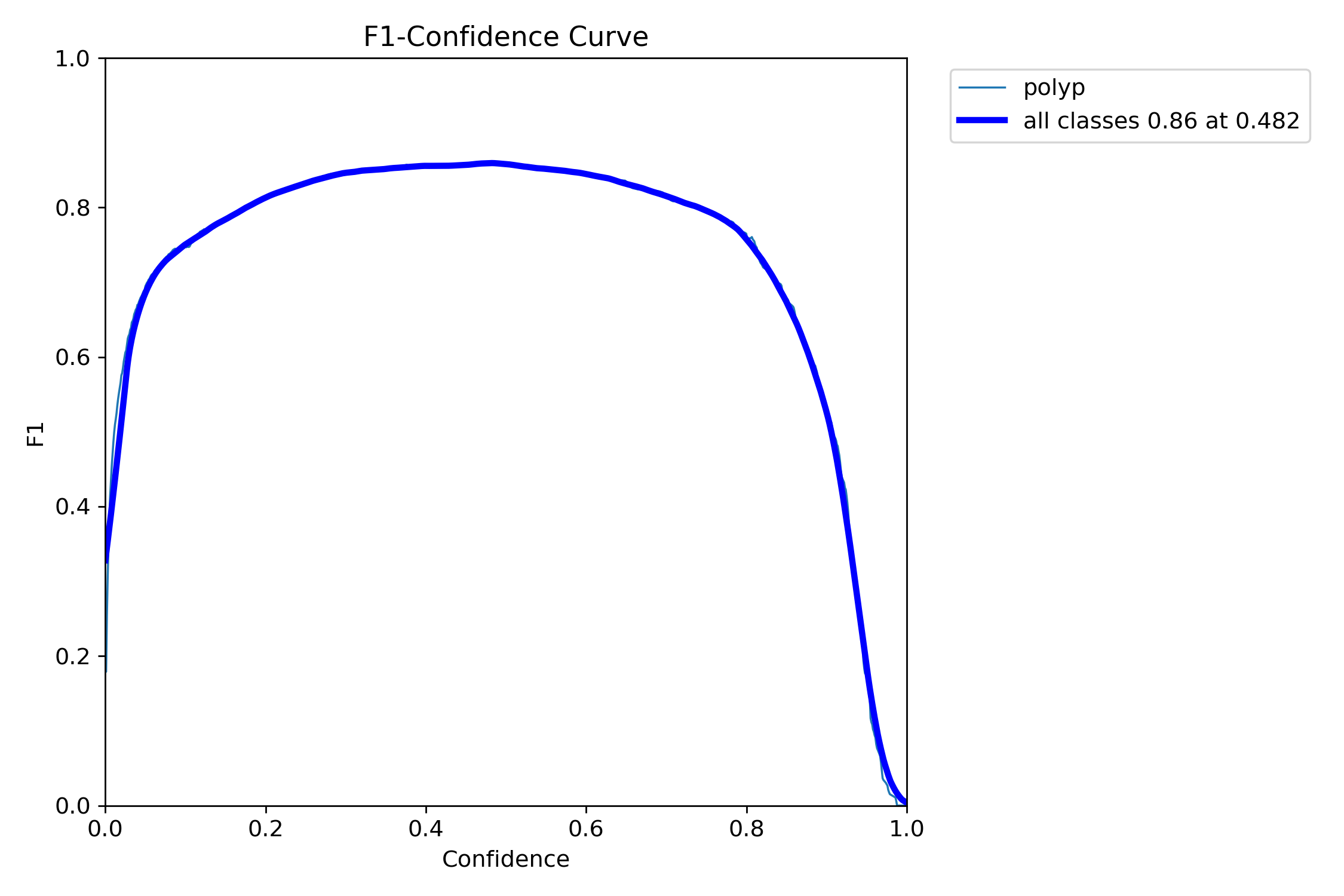
mAP50: 0.9138
mAP50-95: 0.7264
Precision: 0.8923
Recall: 0.8278=== NBI-LCI-FICE-BLI Metrics on unseen modalities===
mAP50: 0.6947
mAP50-95: 0.4972
Precision: 0.8402
Recall: 0.5672This project implements a robust polyp detection system using YOLO11 (You Only Look Once version 11) for medical image analysis. The model is trained on WLI modality.
- Base Model: YOLO11 from Ultralytics
- Input Resolution: 640×640 pixels
- Backbone: CSPDarkNet
- Neck: PANet
- Head: Multi-scale detection
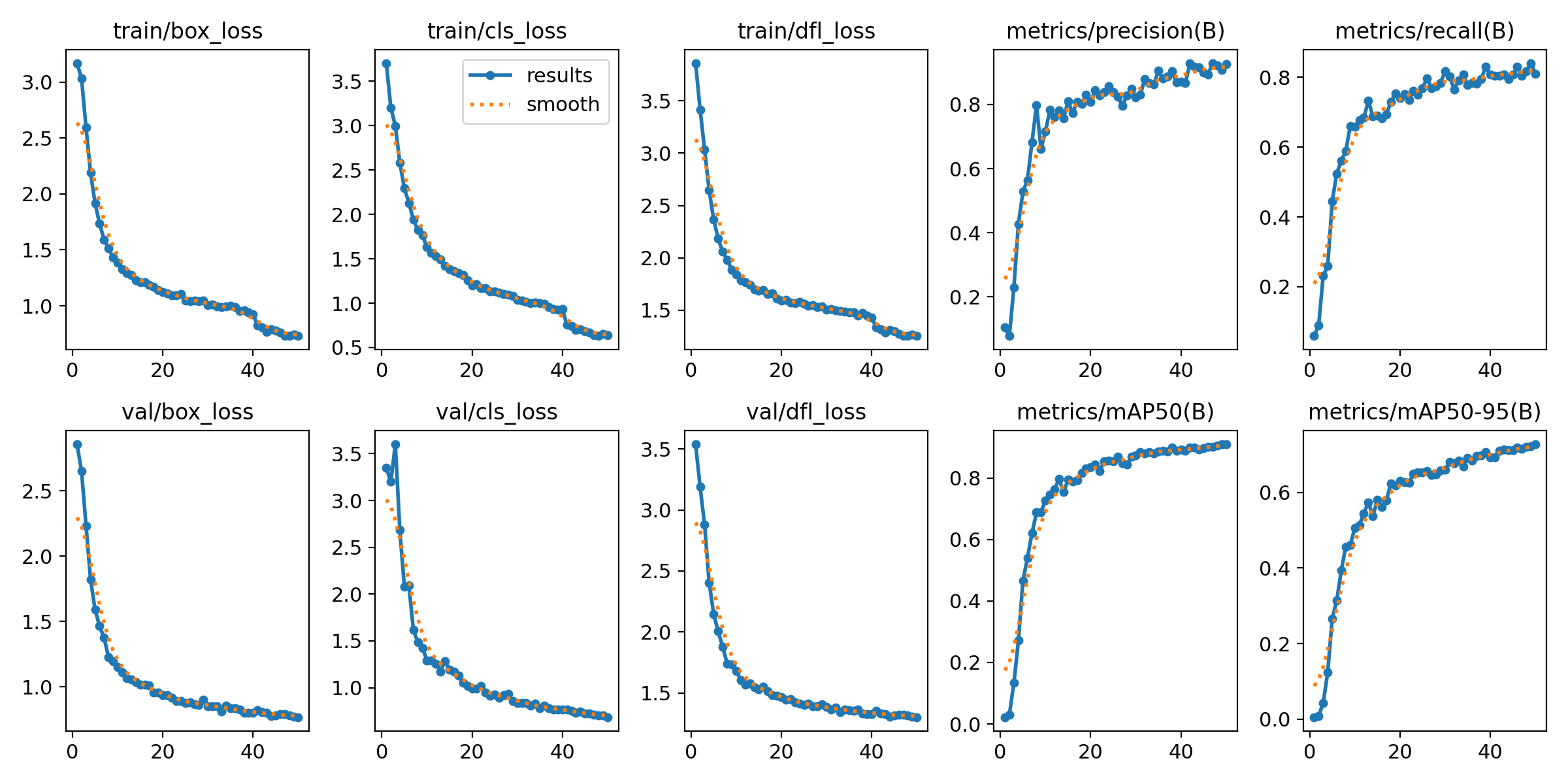
- Epochs: 50
- Batch Size: 16
- Initial Learning Rate: 0.001
- Optimizer: Auto-selected
- Early Stopping Patience: 10 epochs
- Mosaic: 0.8 probability
- MixUp: 0.1 probability
- Copy-Paste: 0.1 probability
- Horizontal Flip: 0.5 probability
- Color Augmentation: HSV adjustments
- Spatial Transformations: Rotation, translation, scaling, shearing
- Training Confidence Threshold: 0.1
- IoU Threshold: 0.4
- Augmentation Focus: Small object detection
=== Test Set Metrics on trained modality(WLI)===
mAP50: 0.9282
mAP50-95: 0.7067
Precision: 0.8799
Recall: 0.8638=== NBI-LCI-FICE-BLI Metrics on unseen modalities===
mAP50: 0.7619
mAP50-95: 0.5383
Precision: 0.8357
Recall: 0.6675=== Metrics on unseen Dataset: REAL-Colon ===
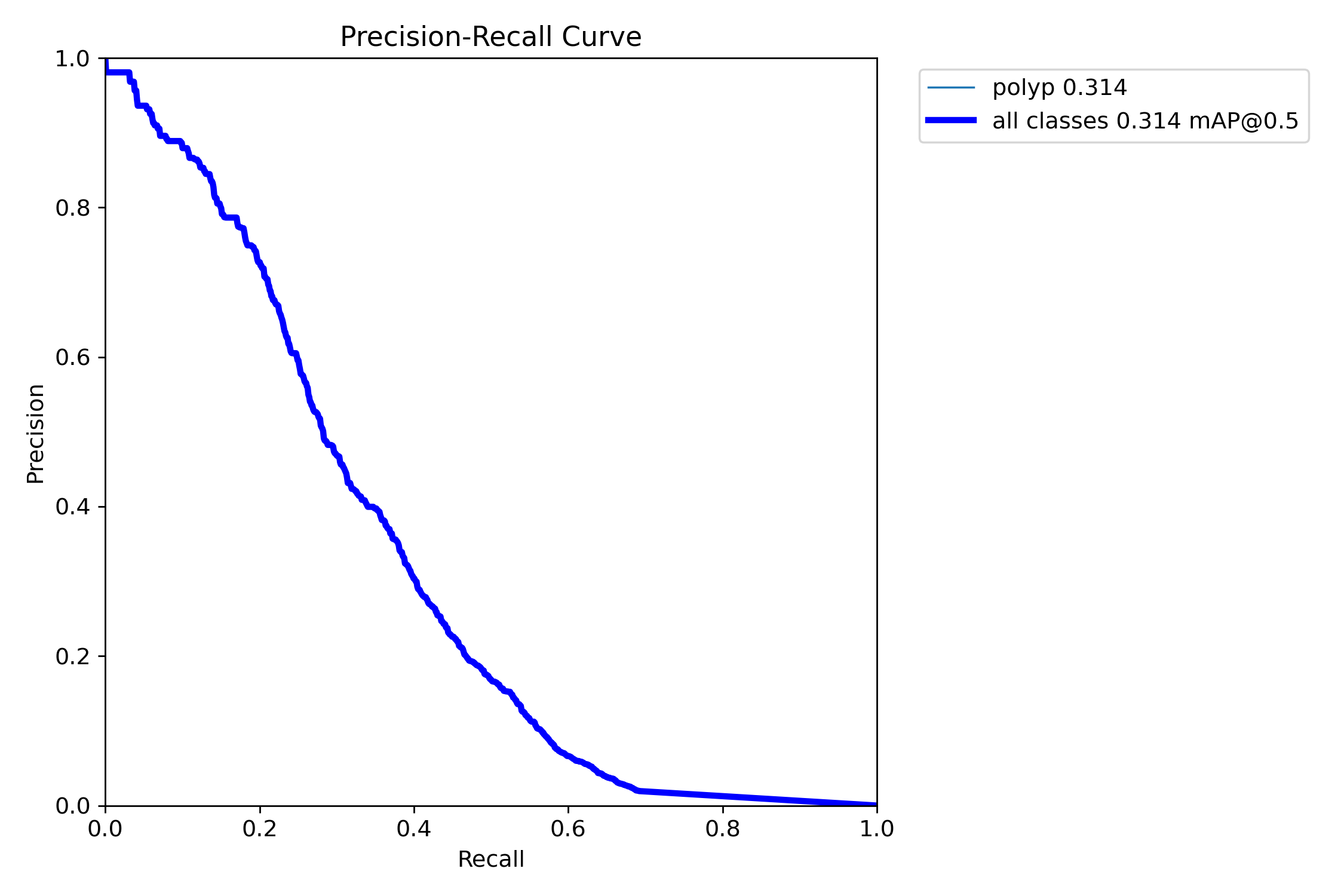
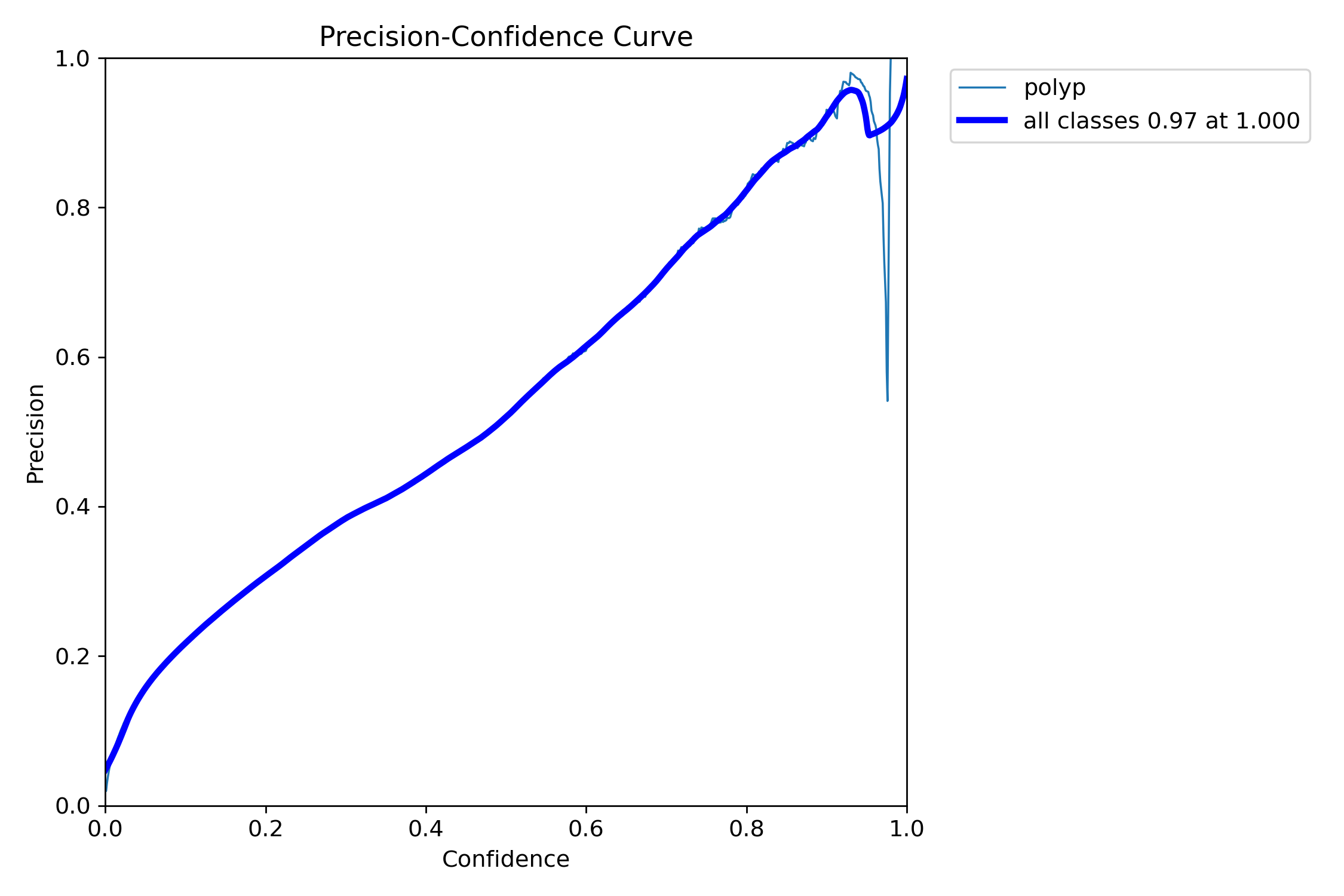
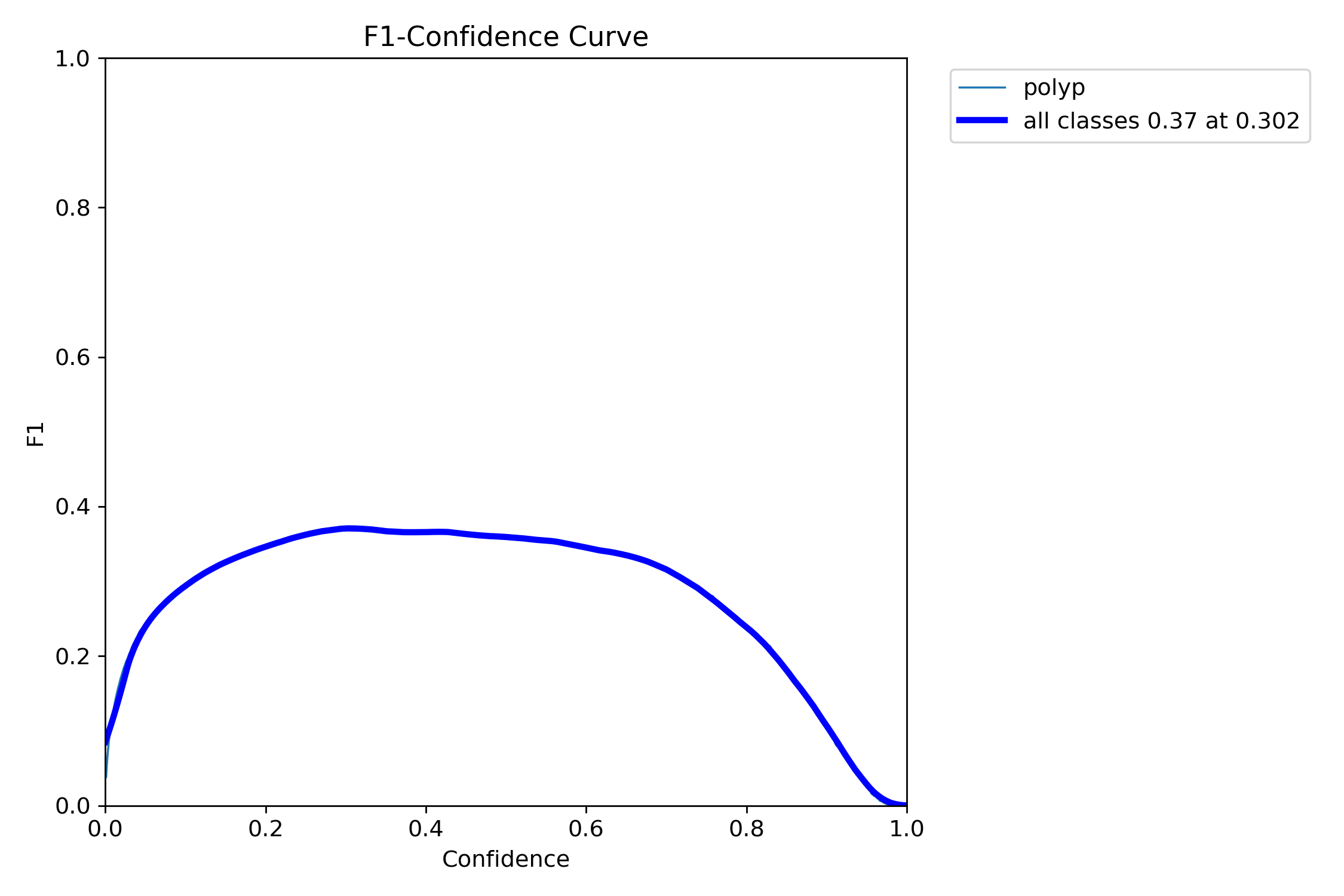
mAP50: 0.3415
mAP50-95: 0.1922
Precision: 0.4986
Recall: 0.3413