All this stuff is collected from the github which was not well maintained. I will soon reorganize this to reflect recency (as a lot of work is happening in comp biology that CNNs are using lately -- hopefully CapsNETs too) and some easy to undrstand structure.
Key collaborators and researchers (besides me) will be some budding computational biologists such as Huadong Liao who did an awesome job with recent Capsule Networks and will work with me to not only do awesome projects across the globe but also work and travel with me to deliver workshops for both for and non-profit organizations.
If you are a researcher, enthusiast or even someone just interested and want to join computational biology researches with deep learning, then contact me at : tarry.singh@gmail.com
So stay tuned!!!
Thanks,
Tarry Singh
This is a list of implementations of deep learning methods to biology, originally published on Follow the Data. There is a slant towards genomics because that's the subfield that I follow most closely.
Please, contribute to this growing list, especially in categories that I haven't covered well! Also, do add your contributions to GitXiv as well if you can.
You might also want to refer to the curaterd list of deepbio work below.
- Data driven decision making
- Questions -> Data -> Models/Tools
- Overview
- EHR data
- Insurance claims data
- Clinical notes
- Image data
- Time series data
- Genomics data
- Deep Learning Computational biology
- Curated List
| Data type | Models/Tools | Applications |
|---|---|---|
| -EHR data -Insurance claims data |
ML(logistic regression,XGBoost) | Predict outcomes (disease, death, readmission etc.) |
| -Clinical notes -Conversation text data |
-Rule based approach(regular expression) -Deep learning apporach |
-Extract concepts from clinical notes -Knowledge graphs -Chat-bot -QA system |
| Medical image data (X-ray, CT, OCR image etc.) | CNN | -Detection: diagnosis of skin cancer lung nodule or diabetic reinopathy -Segmentation of tumor, histopathology |
| Time series data (EEG, ECG, vital sign data etc.) | HMM,RNN,CNN | -Heart disease -Sleep disorder(apnea) -ICU monitoring |
| Genomics data | GATK,QIIME | -Cancer mutation identification -Biomarker identification -Durg discovery |
| Other data (hospital operational data) | -ML(regression) -Queueing model |
-Reduce operational cost -Improve patient experience -ER wait time and queueing |
| Prediction outcomes | Models/Tools | Data type | Sample size | Reference | Year |
|---|---|---|---|---|---|
| Review | Mining electronic health records: towards better research applications and clinical care | 2012 | |||
| Review | Opportunities and challenges in developing risk prediction models with electronic health records data: a systematic review | 2016 | |||
| heart failure | -logistic regression -random forest |
longitudinal EHR data | 1684 heart failure cases and 13525 matched controls | Early Detection of Heart Failure Using Electronic Health Records | 2016 |
| heart failure (review) | Population Risk Prediction Models for Incident Heart Failure | 2015 | |||
| Kidney transplant graft failure | Cox regression | 10-years EHR data | 69,440 kidney transpants | A comprehensive risk quantification score for deceased donor kidneys: the kidney donor risk index | 2009 |
| Prediction outcomes | Models/Tools | Data type | Sample size | Reference | Year |
|---|---|---|---|---|---|
| Review | Realizing the full potential of electronic health records: the role of natural language processing | 2011 | |||
| Review | Natural language processing: an introduction | 2011 | |||
| Negation | Regular expression and rule-based approach | Clinical reports | 2060 discharge summaries | A simple algorithm for identifying negated findings and diseases in discharge summaries | 2001 |
| Using electronic health records to drive discovery in disease genomics | |||||
| NER | discharge summaries | 826 notes | A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries | 2011 |
| Prediction outcomes | Models/Tools | Data type | Sample size | Reference | Year |
|---|---|---|---|---|---|
| Diabetic retinopathy | CNN | retinal fundus images | 128175 retinal images | Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs | 2016 |
| Skin cancer | CNN | skin images | 129,450 skin images | Dermatologist-level classification of skin cancer with deep neural networks | 2017 |
| Tumor | CNN | Pathology images | 400+110 slides | Detecting Cancer Metastases on Gigapixel Pathology Images | 2017 |
| Deep Learning Automates the Quantitative Analysis of Individual Cells in Live-Cell Imaging Experiments |
| Prediction outcomes | Models/Tools | Data type | Sample size | Reference | Year |
|---|---|---|---|---|---|
| sinus rhythm and atrial fibrillation | 34-layer convolutional neural network (CNN) | single-lead ECG | -(Train) 64,121 ECG records from 29,163 patients -(Test) 336 records from 328 unique patients |
Cardiologist-Level Arrhythmia Detection with Convolutional Neural Networks | 2017 |
| Hand movements | CNN | sEMG | 67 intact subjects and 11 transradial amputees | Deep Learning with Convolutional Neural Networks Applied to Electromyography Data: A Resource for the Classification of Movements for Prosthetic Hands | 2016 |
| Review | ICU data | Machine Learning and Decision Support in Critical Care | 2017 |
| Prediction outcomes | Models/Tools | Data type | Sample size | Reference | Year |
|---|---|---|---|---|---|
| Genetic variants | Exome NGS | NGS&EHR data | 50,726 individuals | Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study | 2016 |
| Familial hypercholesterolemia | Exome NGS | NGS&EHR data | 50,726 individuals | Genetic identification of familial hypercholesterolemia within a single U.S. health care system | 2016 |
| Prediction outcomes | Models/Tools | Data type | Sample size | Reference | Year |
|---|---|---|---|---|---|
| Drug discovery | LSTM | Assay | 12-27 assays | Low data drug discovery with one-shot learning | 2017 |
| Tutorial | Image | Deep learning models for health care: challenges and solutions | 2017 | ||
| Tutorial | Image | Deep learning in radiology: recent advances, challenges and future trends | 2016 | ||
| Tutorial | Big data analytics for healthcare | 2013 | |||
| Tutorial | Image | Survey of deep learning in radiology | 2017 | ||
| ER wait time | ER visit time | Accurate ED Wait Time Prediction | 2017 |
This is a list of implementations of deep learning methods to biology, originally published on Follow the Data. There is a slant towards genomics because that's the subfield that I follow most closely.
Please, contribute to this growing list, especially in categories that I haven't covered well! Also, do add your contributions to GitXiv as well if you can.
You might also want to refer to the curaterd list of deepbio work below.
These are not implementations as such, but contain useful pointers.
Opportunities And Obstacles For Deep Learning In Biology And Medicine [bioRxiv preprint]
This impressive collaborative review was written completely in the open on Github. It is focused on discussing how deep learning may be able to transform patient classification and treatment as well as fundamental biological research in the future, and what the main obstacles are that could prevent it from happening. A lot of interesting points are brought up here. Together with the review listed below, which has a more technical slant, you will get a good overview of how deep learning is used and can be used in biology and medicine.
Deep learning for computational biology [open access paper]
This is a very nice review of deep learning applications in biology. It primarily deals with convolutional networks and explains well why and how they are used for sequence (and image) classification.
Deep learning for health informatics [open access paper]
An overview of several types of deep nets and their applications in translational bioinformatics, medical imaging, "pervasive sensing", medical data and public health.
Neural graph fingerprints [github][gitxiv]
A convolutional net that can learn features which are useful for predicting properties of novel molecules; “molecular fingerprints”. The net works on a graph where atoms are nodes and bonds are edges. Developed by the group of Ryan Adams, who co-hosts the very good Talking Machines podcast.
Deep-learning models for Drug Discovery and Quantum Chemistry [github][Python library][preprint]
This is a "... [P]ython library that aims to make the use of machine-learning in drug discovery straightforward and convenient" which checks a lot of boxes when it comes to advanced is deep learning: one-shot learning, graph convolutional networks, learning from less data, and LSTM embeddings. According to the GitHub site, "DeepChem aims to provide a high quality open-source toolchain that democratizes the use of deep-learning in drug discovery, materials science, and quantum chemistry."
Continuous Distributed Representation of Biological Sequences for Deep Genomics and Deep Proteomics[github][paper]
The GitHub summary reads: "We introduce a new representation for biological sequences. Named bio-vectors (BioVec) to refer to biological sequences in general with protein-vectors (ProtVec) for proteins (amino-acid sequences) and gene-vectors (GeneVec) for gene sequences, this representation can be widely used in applications of deep learning in proteomics and genomics. Biovectors are basically n-gram character skip-gram wordvectors for biological sequences (DNA, RNA, and Protein). In this work, we have explored biophysical and biochemical meaning of this space. In addition, in variety of bioinformatics tasks we have shown the strength of such a sequence representation."
Pcons2 – Improved Contact Predictions Using the Recognition of Protein Like Contact Patterns [web interface]
Here, a “deep random forest” with five layers is used to improve predictions of which residues (amino acids) in a protein are physically interacting which each other. This is useful for predicting the overall structure of the protein (a very hard problem.)
This category is divided into several subfields.
In modeling gene expression, the inputs are typically numerical values (integers or floats) estimating how much RNA is produced from a DNA template in a particular cell type or condition.
ADAGE – Analysis using Denoising Autoencoders of Gene Expression [github][gitxiv]
This is a Theano implementation of stacked denoising autoencoders for extracting relevant patterns from large sets of gene expression data, a kind of feature construction approach if you will. I have played around with this package quite a bit myself. The authors initially published a conference paper applying the model to a compendium of breast cancer (microarray) gene expression data, and more recently posted a paper on bioRxiv where they apply it to all available expression data (microarray and RNA-seq) on the pathogen Pseudomonas aeruginosa. (I understand that this manuscript will soon be published in a journal.)
Learning structure in gene expression data using deep architectures [paper]
This is also about using stacked denoising autoencoders for gene expression data, but there is no available implementation (as far as I could tell). Included here for the sake of completeness (or something.)
Gene expression inference with deep learning [github][paper]
This deals with a specific prediction task, namely to predict the expression of specified target genes from a panel of about 1,000 pre-selected “landmark genes”. As the authors explain, gene expression levels are often highly correlated and it may be a cost-effective strategy in some cases to use such panels and then computationally infer the expression of other genes. Based on Pylearn2/Theano.
Learning a hierarchical representation of the yeast transcriptomic machinery using an autoencoder model [paper]
The authors use stacked autoencoders to learn biological features in yeast from thousands of microarrays. They analyze the hidden layer representations and show that these encode biological information in a hierarchical way, so that for instance transcription factors are represented in the first hidden layer.
Here the inputs are typically “raw” DNA sequence, and convolutional networks (or layers) are often used to learn regularities within the sequence. Hat tip to Melissa Gymrek for pointing out some of these.
DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences [github][gitxiv]
Made for predicting the function of non-protein coding DNA sequence. Uses a convolution layer to capture regulatory motifs (i e single DNA snippets that control the expression of genes, for instance), and a recurrent layer (of the LSTM type) to try to discover a “grammar” for how these single motifs work together. Based on Keras/Theano.
Basset – learning the regulatory code of the accessible genome with deep convolutional neural networks [github][gitxiv]
Based on Torch, this package focuses on predicting the accessibility (or “openness”) of the chromatin – the physical packaging of the genetic information (DNA+associated proteins). This can exist in more condensed or relaxed states in different cell types, which is partly influenced by the DNA sequence (not completely, because then it would not differ from cell to cell.)
DeepSEA – Predicting effects of noncoding variants with deep learning–based sequence model [web server][paper]
Like the packages above, this one also models chromatin accessibility as well as the binding of certain proteins (transcription factors) to DNA and the presence of so-called histone marks that are associated with changes in accessibility. This piece of software seems to focus a bit more explicitly than the others on predicting how single-nucleotide mutations affect the chromatin structure. Published in a high-profile journal (Nature Methods).
DeepBind – Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning [code][paper]
This is from the group of Brendan Frey in Toronto, and the authors are also involved in the company Deep Genomics. DeepBind focuses on predicting the binding specificities of DNA-binding or RNA-binding proteins, based on experiments such as ChIP-seq, ChIP-chip, RIP-seq, protein-binding microarrays, and HT-SELEX. Published in a high-profile journal (Nature Biotechnology.)
DeeperBind - Enhancing Prediction of Sequence Specificities of DNA Binding Proteins [preprint]
This is an attempt to improve on DeepBind by adding a recurrent sequence learning module (LSTM) after the convolutional layer(s). In this way, the authors propose to capture a positional dimension that is lost in the pooling step in the original DeepBind design. They claim that benchmarking shows that this architecture leads to superior performance compared to previous work.
DeepMotif - Visualizing Genomic Sequence Classifications [paper]
This is also about learning and predicting binding specificities of proteins to certain DNA patterns or "motifs". However, this paper makes use of a combination of convolutional layers and highway networks, with more layers than the DeepBind network. The authors also show how a learned classifier can generate typical DNA motifs by input optimization; applying back-propagation with all the weights held constant in order to find an input pattern that maximally activates the appropriate output node in the network.
Convolutional Neural Network Architectures for Predicting DNA-Protein Binding [code][paper]
This work describes a systematic exploration of convolutional neural network (CNN) architectures for DNA-protein binding. It concludes that the convolutional kernels are very important for the success of the networks on motif-based tasks. Interestingly, the authors have provided a Dockerized implementation of DeepBind from the Frey lab (see above) and also provide EC2-laucher scripts and code for comparing different GPU enabled models programmed in Caffe.
PEDLA: predicting enhancers with a deep learning-based algorithmic framework [code][paper]
This package is for predicting enhancers (stretches of DNA that can enhance the expression of a gene under certain conditions or in a certain kind of cell, often working at a distance from the gene itself) based on heterogeneous data from (e.g.) the ENCODE project, using 1,114 features altogether.
DEEP: a general computational framework for predicting enhancers [paper][code]
An ensemble prediction method for enhancers.
Genome-Wide Prediction of cis-Regulatory Regions Using Supervised Deep Learning Methods (and several other papers applying various kinds of deep networks to regulatory region prediction) [code] (one [paper] out of several)
Wyeth Wasserman’s group have made a kind of toolkit (based on the Theano tutorials) for applying different kinds of deep learning architectures to cis-regulatory element (DNA stretches that can modulate the expression of a nearby gene) prediction. They use a specific “feature selection layer” in their nets to restrict the number of features in the models. This is implemented as an additional sparse one-to-one linear layer between the input layer and the first hidden layer of a multi-layer perceptron.
FIDDLE: An integrative deep learning framework for functional genomic data inference [paper][code[Youtube talk]
The group predicted transcription start site and regulatory regions but claims this solution could be easily generalized and predict other features too. FIDDLE stands for Flexible Integration of Data with Deep LEarning. The idea (nicely explained by the author in the YouTube video above) is to model several genomic signals jointly using convolutional networks. This could be for example DNase-seq, ATAC-seq, ChIP-seq, TSS-seq, maybe RNA-seq signals (as in .wig files with one value per base in the genome).
DeepLNC, a long non-coding RNA prediction tool using deep neural network [paper] [web server]
Identification of potential long non-coding RNA molecules from DNA sequence, based on k-mer profiles.
DeepCpG - Predicting DNA methylation in single cells [paper] [code] [docs]
DeepCpG is a deep neural network for predicting DNA methylation in multiple cells. DeepCpG has a modular architecture, consisting of a recurrent CpG module to account for correlations between CpG sites within and across cells, a convolutional DNA module to extract patterns from a wide DNA sequence window, and a Joint module that integrates the evidence from the CpG and DNA module to predict the methylation state of multiple cells for a target CpG site. DeepCpG yields accurate predictions, enables discovering DNA sequence motifs that are associated with DNA methylation states and cell-to-cell variability, and can be used for analyzing the effect of single-nucleotide mutations on DNA methylation. DeepCpG is implemented in Python and publicly available.
Predicting DNA Methylation State of CpG Dinucleotide Using Genome Topological Features and Deep Networks [paper][web server]
This implementation uses a stacked autoencoder with a supervised layer on top of it to predict whether a certain type of genomic region called “CpG islands” (stretches with an overrepresentation of a sequence pattern where a C nucleotide is followed by a G) is methylated (a chemical modification to DNA that can modify its function, for instance methylation in the vicinity of a gene is often but not always related to the down-regulation or silencing of that gene.) This paper uses a network structure where the hidden layers in the autoencoder part have a much larger number of nodes than the input layer, so it would have been nice to read the authors’ thoughts on what the hidden layers represent.
DeepCpG - Predicting DNA methylation in single cells [paper] [code] [docs]
See above.
CellCnn – Representation Learning for detection of disease-associated cell subsets [code][paper]
This is a convolutional network (Lasagne/Theano) based approach for “Representation Learning for detection of phenotype-associated cell subsets.” It is interesting because most neural network approaches for high-dimensional molecular measurements (such as those in the gene expression category above) have used autoencoders rather than convolutional nets.
DeepCyTOF: Automated Cell Classification of Mass Cytometry Data by Deep Learning and Domain Adaptation[paper]
Describes autoencoder approaches (stacked AE and multi-AE) to gating (assigning cells into discrete groups) with mass cytometry (CyTOF).
Using Neural Networks To Improve Single-Cell RNA-Seq Data Analysis[preprint]
Tests a variety of neural network architectures for obtaining a reduced representation of single-cell gene expression data. Introduces a database of tens of thousands of single-cell profiles which can be queried to infer a cell type or state based on this reduced representation.
Removal of batch effects using distribution-matching residual networks[code][paper]
Most high-throughput assays in genomics, proteomics etc. are affected to some extent by systematic technical errors, so-called "batch effects". This paper uses a residual neural network to attenuate batch effects by trying to match the distributions of replicate experiments on e.g. single-cell RNA sequencing or mass cytometry.
Deep learning for population genetic inference [code][paper]
Diet networks: thin parameters for fat genomics [manuscript]
This weirdly-named paper addresses the frequently encountered problem in genomics where the number of features is much larger than the number of training examples. Here, it is addressed in the context of SNPs (single-nucleotide polymorphisms, genetic variations between individuals). The authors propose a new network parametrization that reduces the number of free parameters using a multi-task architecture which tries to learn a useful embedding of the input features.
There are potentially lots of implementations that could go here.
Deep learning for neuroimaging: a validation study [paper]
SPINDLE: SPINtronic deep learning engine for large-scale neuromorphic computing [paper]
A curated list of deep learning applications in the field of computational biology
-
2012-07 | Deep architectures for protein contact map prediction | Pietro Di Lena, Ken Nagata and Pierre Baldi Bioinformatics
-
2012-10 | Predicting protein residue–residue contacts using deep networks and boosting | Jesse Eickholt and Jianlin Cheng | Bioinformatics
-
2013-03 | DNdisorder: predicting protein disorder using boosting and deep networks | Jesse Eickholt and Jianlin Cheng | BMC Bioinformatics
-
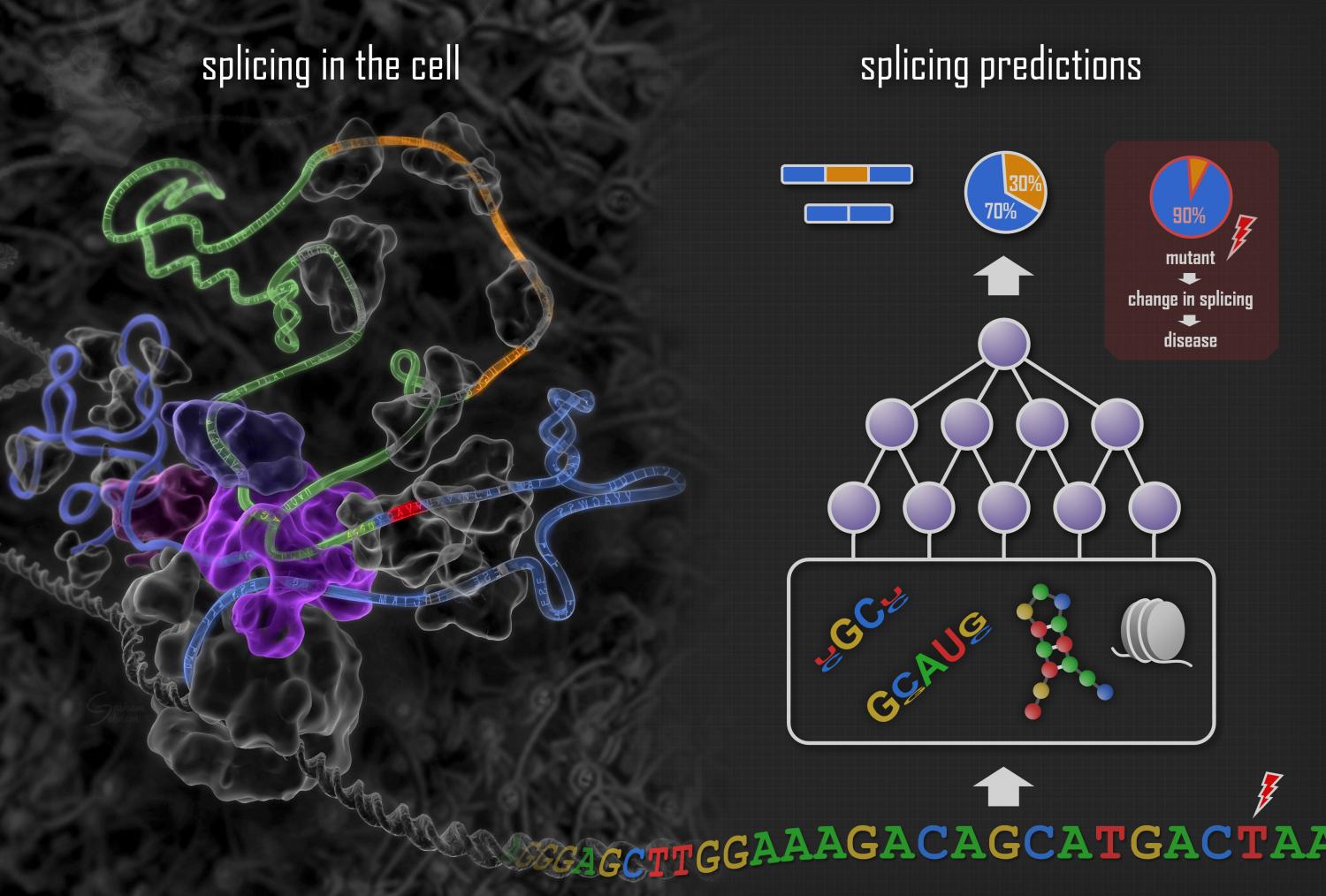
2014-06 | Deep learning of the tissue-regulated splicing code | Michael K. K. Leung, Hui Yuan Xiong, Leo J. Lee and Brendan J. Frey | Bioinformatics
-
2014-10 | DANN: a deep learning approach for annotating the pathogenicity of genetic variants | Daniel Quang, Yifei Chen and Xiaohui Xie | Bioinformatics
-
2014-11 | Pairwise input neural network for target-ligand interaction prediction | Caihua Wang, Juan Liu, Fei Luo, Yafang Tan, Zixin Deng, Qian-Nan Hu | 2014 IEEE International Conference on Bioinformatics and Biomedicine (BIBM)
-
2015-01 | Unsupervised feature construction and knowledge extraction from genome-wide assays of breast cancer with denoising autoencoders. | Jie Tan, Matt Ung, Chao Cheng, Casey Greene | Pacific Symposium on Biocomputing (PSB) | Models & Data
-
2015-01 | The human splicing code reveals new insights into the genetic determinants of disease | Hui Y. Xiong, Babak Alipanahi, Leo J. Lee, Hannes Bretschneider, Daniele Merico, Ryan K. C. Yuen, Yimin Hua, Serge Gueroussov, Hamed S. Najafabadi, Timothy R. Hughes, Quaid Morris, Yoseph Barash, Adrian R. Krainer, Nebojsa Jojic, Stephen W. Scherer, Benjamin J. Blencowe, Brendan J. Frey | Science
-
2015-03 | Deep Feature Selection: Theory and Application to Identify Enhancers and Promoters | Yifeng Li, Chih-Yu Chen, and Wyeth W. Wasserman | 19th Annual International Conference, RECOMB 2015, Warsaw, Proceedings
-
2015-05 | Trans-species learning of cellular signaling systems with bimodal deep belief networks | Lujia Chen, Chunhui Cai, Vicky Chen and Xinghua Lu | Bioinformatics
-
2015-05 | Deep convolutional neural networks for annotating gene expression patterns in the mouse brain | Tao Zeng, Rongjian Li, Ravi Mukkamala, Jieping Ye and Shuiwang Ji | BMC Bioinformatics
-
2015-07 | DeepBind: Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning | Babak Alipanahi, Andrew Delong, Matthew T. Weirauch & Brendan J. Frey | Nature Biotechnology
-
2015-08 | Deep learning for regulatory genomics | Yongjin Park & Manolis Kellis | Nature Biotechnology
-
2015-08 | DeepSEA: Predicting effects of noncoding variants with deep learning–based sequence model | Jian Zhou & Olga G. Troyanskaya | Nature Methods: Short intro & Nature Methods
-
2015-08 | Integrative Data Analysis of Multi-Platform Cancer Data with a Multimodal Deep Learning Approach | Muxuan Liang, Zhizhong Li, Ting Chen, Jianyang Zeng | IEEE/ACM Transactions on Computational Biology and Bioinformatics (TCBB)
-
2015-10 | A deep learning framework for modeling structural features of RNA-binding protein targets | Sai Zhang, Jingtian Zhou, Hailin Hu, Haipeng Gong, Ligong Chen, Chao Cheng, and Jianyang Zeng | NAR
-
2015-10 | Basset: Learning the regulatory code of the accessible genome with deep convolutional neural networks | David R. Kelley, Jasper Snoek, John Rinn | Biorxiv | code
-
2015-10 | Deep Learning for Drug-Induced Liver Injury | Youjun Xu, Ziwei Dai, Fangjin Chen, Shuaishi Gao, Jianfeng Pei, and Luhua Lai | ASC Journal of Chemical Information and Modeling
-
2016-01 | ADAGE-Based Integration of Publicly Available Pseudomonas aeruginosa Gene Expression Data with Denoising Autoencoders Illuminates Microbe-Host Interactions | mSystems | code
-
2015-11 | De novo identification of replication-timing domains in the human genome by deep learning | Feng Liu, Chao Ren, Hao Li, Pingkun Zhou, Xiaochen Bo and Wenjie Shu | Bioinformatics
-
2015-11 | Recurrent Neural Network Based Hybrid Model of Gene Regulatory Network | Khalid Raza, Mansaf Alam | Arxiv
-
2015-11 | Continuous Distributed Representation of Biological Sequences for Deep Proteomics and Genomics | Ehsaneddin Asgari, Mohammad R. K. Mofrad | PloS one
-
2016-01 | Learning a hierarchical representation of the yeast transcriptomic machinery using an autoencoder model | Lujia Chen, Chunhui Cai, Vicky Chen and Xinghua Lu | BMC Bioinformatics
-
2016-01 | PEDLA: predicting enhancers with a deep learning-based algorithmic framework | Feng Liu, Hao Li, Chao Ren, Xiaochen Bo, Wenjie Shu | Biorxiv
-
2016-01 | TensorFlow: Biology’s Gateway to Deep Learning? | Ladislav Rampasek, Anna Goldenberg | Cell Systems
-
2016-01 | ADAGE-Based Integration of Publicly Available Pseudomonas aeruginosa Gene Expression Data with Denoising Autoencoders Illuminates Microbe-Host Interactions | mSystems | code
-
2016-01 | Deep Learning in Drug Discovery | Erik Gawehn, Jan A. Hiss and Gisbert Schneider | Molecular Informatics
-
2016-02 | Gene expression inference with deep learning | Yifei Chen, Yi Li, Rajiv Narayan, Aravind Subramanian, Xiaohui Xie | Bioinformatics
-
2016-02 | Semi-Supervised Learning of the Electronic Health Record for Phenotype Stratification | Brett Beaulieu-Jones, Casey Greene | bioRxiv
-
2016-03 | Genome-Wide Prediction of cis-Regulatory Regions Using Supervised Deep Learning Methods | Yifeng Li, Wenqiang Shi, Wyeth W Wasserman | Biorxiv
-
2016-03 | Applications of deep learning in biomedicine | Polina Mamoshina, Armando Vieira, Evgeny Putin, and Alex Zhavoronkov | ACS Molecular Pharmaceutics
-
2016-03 | Deep Learning in Bioinformatics | Seonwoo Min, Byunghan Lee, Sungroh Yoon | Arxiv
-
2016-03 | DeepNano: Deep Recurrent Neural Networks for Base Calling in MinION Nanopore Reads | Vladimír Boža, Broňa Brejová, Tomáš Vinař | Arxiv | code
-
2016-03 | deepTarget: End-to-end Learning Framework for microRNA Target Prediction using Deep Recurrent Neural Networks | Byunghan Lee, Junghwan Baek, Seunghyun Park, Sungroh Yoon | Arxiv
-
2016-03 | Deep Learning in Label-free Cell Classification | Claire Lifan Chen, Ata Mahjoubfar, Li-Chia Tai, Ian K. Blaby, Allen Huang, Kayvan Reza Niazi & Bahram Jalali | Nature Scientific Reports
-
2016-04 | Accurate classification of protein subcellular localization from high throughput microscopy images using deep learning | Tanel Pärnamaa, Leopold Parts | bioRxiv
-
2016-04 | DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences | Daniel Quang & Xiaohui Xie | Nucleic Acids Research | code
-
2016-04 | deepMiRGene: Deep Neural Network based Precursor microRNA Prediction | Seunghyun Park, Seonwoo Min, Hyun-soo Choi, and Sungroh Yoon | Arxiv
-
2016-04 | Microscopy cell counting and detection with fully convolutional regression networks | Weidi Xie, J. Alison Noble and Andrew Zisserman | Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization
-
2016-04 | Protein Secondary Structure Prediction Using Cascaded Convolutional and Recurrent Neural Networks | Zhen Li and Yizhou Yu | Arxiv
-
2016-05 | Denoising genome-wide histone ChIP-seq with convolutional neural networks | Pang Wei Koh, Emma Pierson, Anshul Kundaje | Biorxiv
-
2016-05 | Deep Motif: Visualizing Genomic Sequence Classifications | Jack Lanchantin, Ritambhara Singh, Zeming Lin, Yanjun Qi | Arxiv
-
2016-05 | Not Just a Black Box: Learning Important Features Through Propagating Activation Differences | Avanti Shrikumar, Peyton Greenside, Anna Shcherbina, Anshul Kundaje | Arxiv
-
2016-05 | Deep biomarkers of human aging: Application of deep neural networks to biomarker development | Evgeny Putin, Polina Mamoshina, Alexander Aliper, Mikhail Korzinkin, Alexey Moskalev, Alexey Kolosov, Alexander Ostrovskiy, Charles Cantor, Jan Vijg, and Alex Zhavoronkov | Aging
-
2016-05 | Deep learning applications for predicting pharmacological properties of drugs and drug repurposing using transcriptomic data | Alexander Aliper, Sergey Plis, Artem Artemov, Alvaro Ulloa, Polina Mamoshina, and Alex Zhavoronkov | ACS Molecular Pharmaceutics
-
2016-05 | Accurate prediction of single-cell DNA methylation states using deep learning | Christof Angermueller, Heather Lee, Wolf Reik, Oliver Stegle | Biorxiv
-
2016-05 | Deep Machine Learning provides state-of-the-art performance in image-based plant phenotyping | Michael P. Pound, Alexandra J. Burgess, Michael H. Wilson, Jonathan A. Atkinson, Marcus Griffiths, Aaron S. Jackson, Adrian Bulat, Yorgos Tzimiropoulos, Darren M. Wells, Erik H. Murchie, Tony P. Pridmore, Andrew P. French | Biorxiv
-
2016-05 | Genetic Architect: Discovering Genomic Structure with Learned Neural Architectures | Laura Deming, Sasha Targ, Nate Sauder, Diogo Almeida, Chun Jimmie Ye | Arxiv
-
2016-05 | DeepCyTOF: Automated Cell Classification of Mass Cytometry Data by Deep Learning and Domain Adaptation | Huamin Li, Uri Shaham, Yi Yao, Ruth Montgomery, Yuval Kluger | Biorxiv
-
2016-06 | Classifying and segmenting microscopy images with deep multiple instance learning | Oren Z. Kraus, Jimmy Lei Ba and Brendan J. Frey | Bioinformatics
-
2016-06 | Convolutional neural network architectures for predicting DNA–protein binding | Haoyang Zeng, Matthew D. Edwards, Ge Liu and David K. Gifford | Bioinformatics | code
-
2016-06 | DeepLNC, a long non-coding RNA prediction tool using deep neural network | Rashmi Tripathi, Sunil Patel, Vandana Kumari, Pavan Chakraborty, Pritish Kumar Varadwaj | Network Modeling Analysis in Health Informatics and Bioinformatics
-
2016-06 | Virtual Screening: A Challenge for Deep Learning | Javier Pérez-Sianes, Horacio Pérez-Sánchez, Fernando Díaz | 10th International Conference on Practical Applications of Computational Biology & Bioinformatics
-
2016-07 | Deep learning for computational biology | Christof Angermueller, Tanel Pärnamaa, Leopold Parts, Oliver Stegle | Molecular Systems Biology
-
2016-07 | Deep Learning in Bioinformatics | Seonwoo Min, Byunghan Lee, Sungroh Yoon | Briefings in Bioinformatics
-
2016-08 | DeepChrome: deep-learning for predicting gene expression from histone modifications | Ritambhara Singh, Jack Lanchantin, Gabriel Robins, Yanjun Qi | Bioinformatics
-
2016-08 | Deep Artificial Neural Networks and Neuromorphic Chips for Big Data Analysis: Pharmaceutical and Bioinformatics Applications | Lucas Antón Pastur-Romay, Francisco Cedrón, Alejandro Pazos and Ana Belén Porto-Pazos | International Journal of Molecular Sciences
-
2016-08 | Deep GDashboard: Visualizing and Understanding Genomic Sequences Using Deep Neural Networks | Jack Lanchantin, Ritambhara Singh, Beilun Wang, Yanjun Qi | Arxiv
-
2016-08 | Modeling translation elongation dynamics by deep learning reveals new insights into the landscape of ribosome stalling | Sai Zhang, Hailin Hu, Jingtian Zhou, Xuan He and Jianyang Zeng | bioRxiv
-
2016-08 | DeepWAS: Directly integrating regulatory information into GWAS using deep learning supports master regulator MEF2C as risk factor for major depressive disorder | Gökcen Eraslan, Janine Arloth, Jade Martins, Stella Iurato, Darina Czamara, Elisabeth B. Binder, Fabian J. Theis, Nikola S. Mueller | bioRxiv
-
2016-09 | The Next Era: Deep Learning in Pharmaceutical Research | Sean Ekins | Pharmaceutical Research
-
2016-09 | Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model | Sheng Wang, Siqi Sun, Zhen Li, Renyu Zhang, Jinbo Xu | Arxiv
-
2016-10 | Automatic chemical design using a data-driven continuous representation of molecules | Rafael Gómez-Bombarelli, David Duvenaud, José Miguel Hernández-Lobato, Jorge Aguilera-Iparraguirre, Timothy D. Hirzel, Ryan P. Adams, Alán Aspuru-Guzik | Arxiv
-
2016-10 | FIDDLE: An integrative deep learning framework for functional genomic data inference | Umut Eser, L. Stirling Churchman | bioRxiv
-
2016-10 | Deep Learning for Imaging Flow Cytometry: Cell Cycle Analysis of Jurkat Cells | Philipp Eulenberg, Niklas Koehler, Thomas Blasi, Andrew Filby, Anne E. Carpenter, Paul Rees, Fabian J. Theis, F. Alexander Wolf | bioRxiv
-
2016-10 | Leveraging uncertainty information from deep neural networks for disease detection | Christian Leibig, Vaneeda Allken, Philipp Berens, Siegfried Wahl | bioRxiv
-
2016-11 | Predicting Enhancer-Promoter Interaction from Genomic Sequence with Deep Neural Networks | Shashank Singh, Yang Yang, Barnabas Poczos, Jian Ma | bioRxiv
-
2016-11 | RNA-protein binding motifs mining with a new hybrid deep learning based cross-domain knowledge integration approach | Xiaoyong Pan, Hong-Bin Shen | bioRxiv
-
2016-11 | Low Data Drug Discovery with One-shot Learning | Han Altae-Tran, Bharath Ramsundar, Aneesh S. Pappu, Vijay Pande | Arxiv
-
2016-11 | Diet Networks: Thin Parameters for Fat Genomic | Adriana Romero, Pierre Luc Carrier, Akram Erraqabi, Tristan Sylvain, Alex Auvolat, Etienne Dejoie, Marc-André Legault, Marie-Pierre Dubé, Julie G. Hussin, Yoshua Bengio | Arxiv
-
2016-11 | DeeperBind: Enhancing Prediction of Sequence Specificities of DNA Binding Proteins | Hamid Reza Hassanzadeh, May D. Wang | Arxiv
-
2016-11 | Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model | Sheng Wang, Siqi Sun, Zhen Li, Renyu Zhang, Jinbo Xu | bioRxiv
-
2016-11 | Deep learning with feature embedding for compound-protein interaction prediction | Fangping Wan, Jianyang Zeng | bioRxiv
-
2016-12 | Creating a universal SNP and small indel variant caller with deep neural networks | Ryan Poplin, Dan Newburger, Jojo Dijamco, Nam Nguyen, Dion Loy, Sam S. Gross, Cory Y. McLean, Mark A. DePristo | bioRxiv
-
2016-12 | DeepCancer: Detecting Cancer through Gene Expressions via Deep Generative Learning | Rajendra Rana Bhat, Vivek Viswanath, Xiaolin Li | Arxiv
-
2016-12 | Cox-nnet: an artificial neural network Cox regression for prognosis prediction | Travers Ching, Xun Zhu, Lana Garmire | bioRxiv
-
2016-12 | Deep learning is effective for the classification of OCT images of normal versus Age-related Macular Degeneration | Cecilia S Lee, Doug M Baughman, Aaron Y Lee | bioRxiv
-
2016-12 | Partitioned learning of deep Boltzmann machines for SNP data | Moritz Hess, Stefan Lenz, Tamara Blaette, Lars Bullinger, Harald Binder | bioRxiv
-
2016-12 | DeepAD: Alzheimer′s Disease Classification via Deep Convolutional Neural Networks using MRI and fMRI | Saman Sarraf, John Anderson, Ghassem Tofighi, for the Alzheimer's Disease Neuroimaging Initiativ | bioRxiv
-
2016-12 | Training Genotype Callers with Neural Networks | Rémi Torracinta, Fabien Campagne | bioRxiv
-
2016-12 | EP-DNN: A Deep Neural Network-Based Global Enhancer Prediction Algorithm | Seong Gon Kim, Mrudul Harwani, Ananth Grama, Somali Chaterji | Nature Scientific Reports
-
2016-12 | EnhancerPred: a predictor for discovering enhancers based on the combination and selection of multiple features | Cangzhi Jia, Wenying He | Nature Scientific Reports
-
2016-12 | DeepEnhancer: Predicting enhancers by convolutional neural networks | Min, Xu, Ning Chen, Ting Chen, and Rui Jiang | 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM)
-
2016-12 | DeepSplice: Deep classification of novel splice junctions revealed by RNA-seq | Zhang, Yi, Xinan Liu, James N. MacLeod, and Jinze Liu | 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM)
-
2016-12 | Deep convolutional neural networks for detecting secondary structures in protein density maps from cryo-electron microscopy | Li, Rongjian, Dong Si, Tao Zeng, Shuiwang Ji, and Jing He | 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM)
-
2016-12 | Towards recognition of protein function based on its structure using deep convolutional networks | Tavanaei, Amirhossein, Anthony S. Maida, Arun Kaniymattam, and Rasiah Loganantharaj | 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM)
-
2016-12 | Emotion recognition from multi-channel EEG data through Convolutional Recurrent Neural Network | Li, Xiang, Dawei Song, Peng Zhang, Guangliang Yu, Yuexian Hou, and Bin Hu | 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM)
-
2016-12 | Coarse-to-Fine Stacked Fully Convolutional Nets for lymph node segmentation in ultrasound images | Zhang, Yizhe, Michael TC Ying, Lin Yang, Anil T. Ahuja, and Danny Z. Chen | 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM)
-
2016-12 | CNNsite: Prediction of DNA-binding residues in proteins using Convolutional Neural Network with sequence features | Zhou, Jiyun, Qin Lu, Ruifeng Xu, Lin Gui, and Hongpeng Wang | 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM)
-
2016-12 | A predictive model of gene expression using a deep learning framework | Xie, Rui, Andrew Quitadamo, Jianlin Cheng, and Xinghua Shi | 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM)
-
2016-12 | Deep convolutional neural network for survival analysis with pathological images | Zhu, Xinliang, Jiawen Yao, and Junzhou Huang | 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM)
-
2016-12 | Dependency-based convolutional neural network for drug-drug interaction extraction | Liu, Shengyu, Kai Chen, Qingcai Chen, and Buzhou Tang | 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM)
-
2016-12 | Pervasive EEG diagnosis of depression using Deep Belief Network with three-electrodes EEG collector | Cai, Hanshu, Xiaocong Sha, Xue Han, Shixin Wei, and Bin Hu | 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM)
-
2016-12 | Cardiac left ventricular volumes prediction method based on atlas location and deep learning | Luo, Gongning, Suyu Dong, Kuanquan Wang, and Henggui Zhang | 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM)
-
2016-12 | A high-precision shallow Convolutional Neural Network based strategy for the detection of Genomic Deletions | Wang, Jing, Cheng Ling, and Jingyang Gao | 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM)
-
2016-12 | The cornucopia of meaningful leads: Applying deep adversarial autoencoders for new molecule development in oncology | Kadurin, Artur, Alexander Aliper, Andrey Kazennov, Polina Mamoshina, Quentin Vanhaelen, Kuzma Khrabrov, and Alex Zhavoronkov | Oncotarget
-
2016-12 | Medical Image Synthesis with Context-Aware Generative Adversarial Networks | Dong Nie, Roger Trullo, Caroline Petitjean, Su Ruan, Dinggang Shen | Arxiv
-
2016-12 | Unsupervised Learning from Noisy Networks with Applications to Hi-C Data | Wang, Bo, Junjie Zhu, Armin Pourshafeie, Oana Ursu, Serafim Batzoglou, and Anshul Kundaje | Advances in Neural Information Processing Systems (NIPS 2016)
-
2016-12 | Deep Learning for Health Informatics | Daniele Ravì, Charence Wong, Fani Deligianni, Melissa Berthelot, Javier Andreu-Perez, Benny Lo, and Guang-Zhong Yang | IEEE Journal of Biomedical and Health Informatics
-
2017-01 | A Deep Learning Approach for Cancer Detection and Relevant Gene Identification | Wang, Jing, Cheng Ling, and Jingyang Gao | Pacific Symposium on Biocomputing 2017
-
2017-01 | Deep Motif Dashboard: Visualizing and Understanding Genomic Sequences Using Deep Neural Networks | Lanchantin, Jack, Ritambhara Singh, Beilun Wang, and Yanjun Qi | Pacific Symposium on Biocomputing 2017
-
2017-01 | HLA class I binding prediction via convolutional neural networks | Yeeleng Scott Vang, Xiaohui Xie | bioRxiv
-
2017-01 | DeadNet: Identifying Phototoxicity from Label-free Microscopy Images of Cells using Deep ConvNets | David Richmond, Anna Payne-Tobin Jost, Talley Lambert, Jennifer Waters, Hunter Elliott | arXiv
-
2017-01 | Dermatologist-level classification of skin cancer with deep neural networks | Andre Esteva, Brett Kuprel, Roberto A. Novoa, Justin Ko, Susan M. Swetter, Helen M. Blau & Sebastian Thrun | Nature
-
2017-01 | Understanding sequence conservation with deep learning | Yi Li, Daniel Quang, Xiaohui Xie | Biorxiv
-
2017-01 | Learning the Structural Vocabulary of a Network | Saket Navlakha | Neural Computation
-
2017-01 | Mining the Unknown: Assigning Function to Noncoding Single Nucleotide Polymorphisms | Sierra S. Nishizaki, Alan P. Boyle | Trends in Genetics
-
2017-01 | Reverse-complement parameter sharing improves deep learning models for genomics | Avanti Shrikumar, Peyton Greenside, Anshul Kundaje | bioRxiv
-
2017-01 | TIDE: predicting translation initiation sites by deep learning | Sai Zhang, Hailin Hu, Tao Jiang, Lei Zhang, Jianyang Zeng | bioRxiv
-
2017-01 | Integrative Deep Models for Alternative Splicing | Anupama Jha, Matthew R Gazzara, Yoseph Barash | bioRxiv
-
2017-01 | Deep Recurrent Neural Network for Protein Function Prediction from Sequence | Xueliang Leon Liu | bioRxiv
-
2017-01 | Nucleotide sequence and DNaseI sensitivity are predictive of 3D chromatin architecture | Jacob Schreiber, Maxwell Libbrecht, Jeffrey Bilmes, William Noble | bioRxiv
-
2017-02 | Imputation for transcription factor binding predictions based on deep learning | Qian Qin, Jianxing Feng | PloS Computational Biology
-
2017-02 | Deep Learning based multi-omics integration robustly predicts survival in liver cancer | Kumardeep Chaudhary, Olivier B. Poirion, Liangqun Lu, Lana Garmire | bioRxiv
-
2017-03 | Predicting the impact of non-coding variants on DNA methylation | Zeng, Haoyang, and David K. Gifford | Nucleic Acids Research
-
2017-03 | H&E-stained Whole Slide Image Deep Learning Predicts SPOP Mutation State in Prostate Cancer | Andrew J Schaumberg, Mark A Rubin, Thomas J Fuchs | bioRxiv
Feel free to send a pull request.