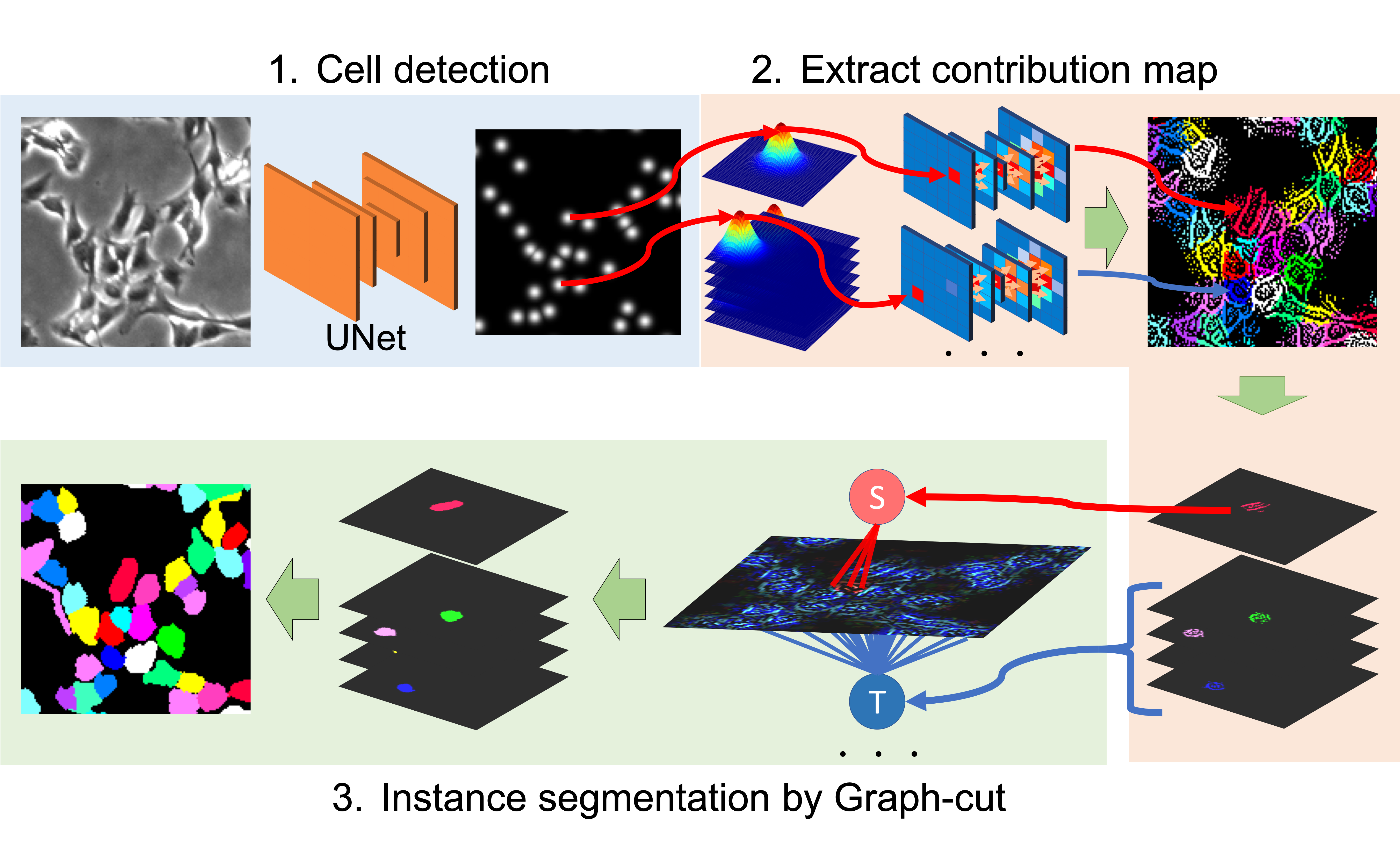
by Kazuya Nishimura, Ker Dai Fei Elmer, Ryoma Bise
- python >= 3.6
- ubuntu 18.04
- CPU or GPU(NVIDIA Driver >= 430)
- matlab
Python setting
conda env create -f=env_name.yml
conda activate pytorchdocker build ./docker
sh run_docker.sh
Graph-cut setting
We use following code.
https://jp.mathworks.com/matlabcentral/fileexchange/38555-kernel-graph-cut-image-segmentation
mkdir graphcut
cd graphcut
wget http://www.wisdom.weizmann.ac.il/~bagon/matlab_code/GCmex1.9.tar.gz
tar -zxvf GCmex1.9.tar.gz
matlab -nodesktop -nosplash -r 'compile_gc; exit'
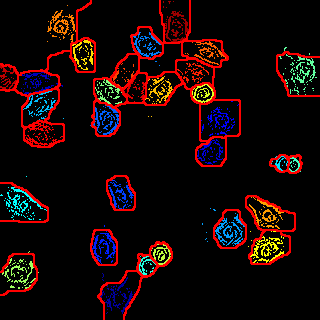
cd ..This demo is only one image's demo. If you want to apply this method to your dataset, you should prepare the likelihood map.
python propagate_main.py -gpython detection_train.py -i :input path(str)
-o :output path(str)
-w :weight path want to load
-g :whether use CUDA
matlab -nodesktop -nosplash -r 'graphcut; exit'This is a sample code.
We don't provide dataset.
If you want to apply your dataset, you should prepare the original image and point level annotation(cell centroid). The attached text file (sample_cell_position.txt) contains a cell position(frame,x,y) as each row. Prepare the same format text file for your dataset.
Set the variance to a value sufficiently larger than the target object. The guided backpropagation depends on variance size.
python likelymapgen.py -i :txt_file_path (str)
-o :output_path (str)
-w :width (int)
-h :height (int)
-g :gaussian variance size (int)
python detection_train.py -gpython detection_train.py -t :train path(str)
-v :validation path(str)
-w :save path of weight(str)
-g :whether use CUDA
-b :batch size (default is 16)
-e :epochs (default is 500)
-l :learning rate(default is 1e-3)
python detection_train.py -gpython detection_train.py -i :input path(str)
-o :output path(str)
-w :weight path want to load
-g :whether use CUDA
If you find the code useful for your research, please cite:
@inproceedings{nishimura2019weakly,
title={Weakly Supervised Cell Instance Segmentation by Propagating from Detection Response},
author={Nishimura, Kazuya and Bise, Ryoma and others},
booktitle={International Conference on Medical Image Computing and Computer-Assisted Intervention},
pages={649--657},
year={2019},
organization={Springer}
}