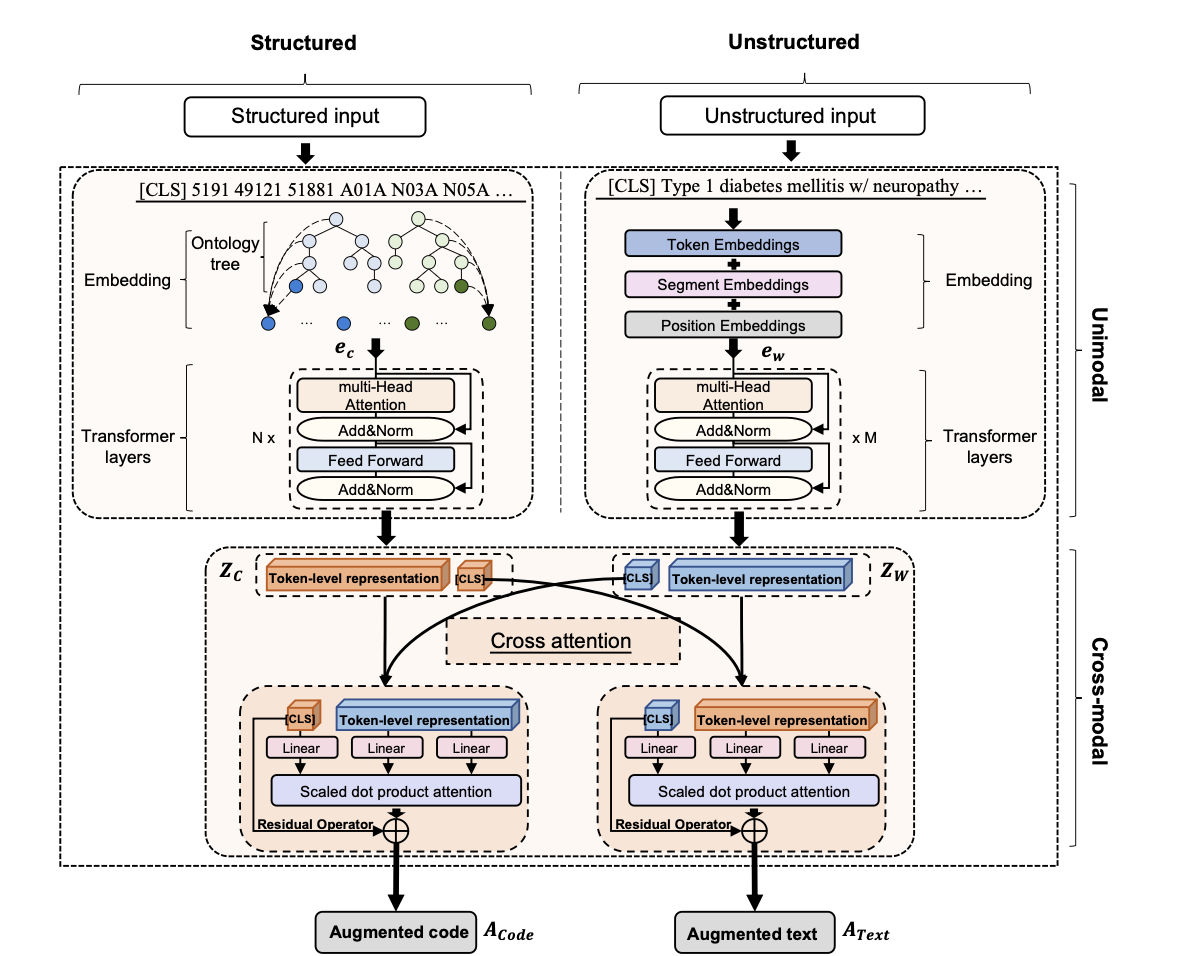
This repository provides the code for Multimodal data matters: language model pre-training over structured and unstructured electronic health records
MedM-PLM handles the inputs from both unstructured and structured EHRs and fuses the information of two modalities in a unified manner. The architecture of MedM-PLM:
To reproduce the steps necessary for pre-training MedM-PLM, you can use the script at the pretrain/run_main.sh
The clinical notes pre-trained model download from ClinicalBERT
CUDA_VISIBLE_DEVICES=0,1 python main.py \
--model_name multi_modal_coAtt_residual_pretrain \
--num_train_epochs 200 \
--do_train \
--graph
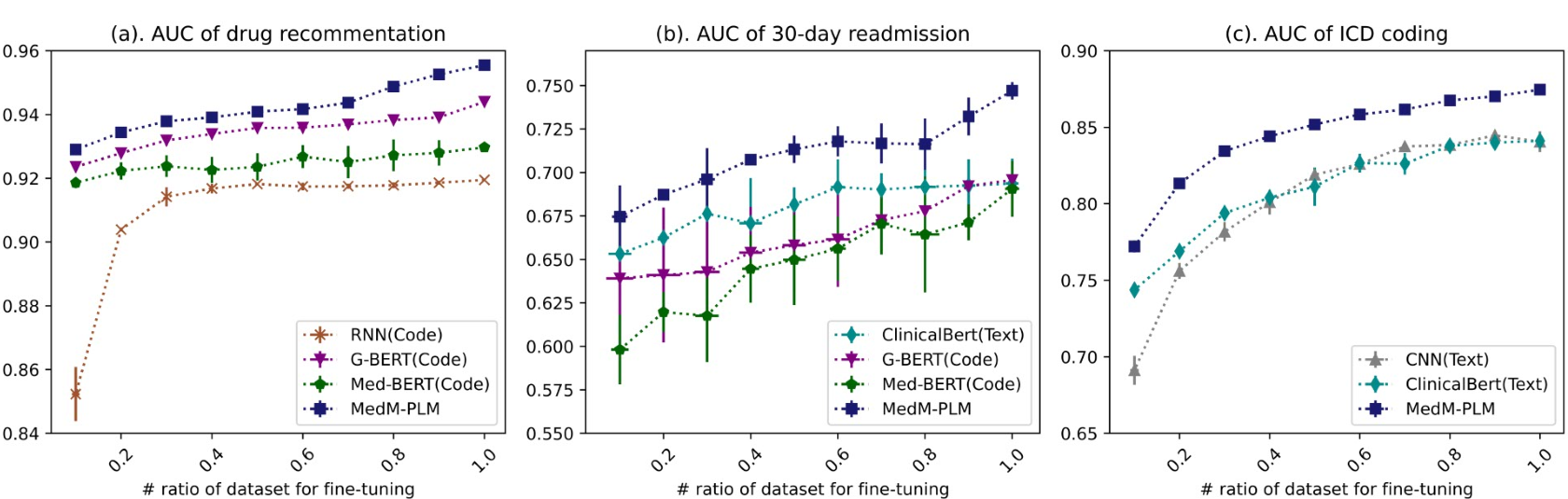
We have conducted three fine-tuning task:
-
Medication recommendation Mediction recommendation is an important application in healthcare, which aims to build computational models to automatically recommend medications that are suited for a patient's health condition. This task is defined as a multi-label prediction issue. For code-aspect, we concatenated the meam of augmented representations of diagnosis and medications in the historical records(0->t-1th visits) and the augmented representation of diagnoses for the t-th visit, while for the text-aspect, we concatenated the mean of fused Text-to-Code representation of historical records to predic the drug of the t-th visit.
CUDA_VISIBLE_DEVICES=0,1 python fusionBert_predict.py \ --learning_rate 5e-5 \ --seed 2022 \ --model_name FusionBert_coAtt_residual_rx_predict \ --graph \ --do_train \ --data_name multilabel \ --predict_task rx \ --model_choice fusion_ml \ --use_pretrain \ --pretrain_dir $pretrained_model_path -
ICD coding
ICD coding fro large-scale clinical notes is labor intensive and error prone. ICD coding usually is treated a multi-label classification problem.
The ICD coding dataset is based on the caml-mimic
-
30-day readmission The task considers a patient encounters a "readmission" if the admission date of patient was within 30 days after the discharge date of the previous hospitalization, and was treated as a binary classification task.
CUDA_VISIBLE_DEVICES=0,1 python fusionBert_predict.py \ --learning_rate 2e-5 \ --seed 2022 \ --model_name FusionBert_coAtt_residual_rx_predict \ --graph \ --do_train \ --data_name binarylabel \ --predict_task readm \ --model_choice fusion_bin \ --use_pretrain \ --pretrain_dir $pretrained_model_path
The details of fine-tuning can find in the folder "finetuning".
We provide script for preprocessing EHRs on MIMIC-III.
The details can find in the folder dataset.
The pre-training dataset MIMIC-III 1.4 is publicly available at https://mimic.physionet.org/. This is a restricted-accessed resource hosted by PhysioNet, which should be used under the license from the credentialed account of PhysioNet.
- Download MIMIC-III data and put in ./dataset.
- There are six tables that need to be downloaded.:
- PRESCRIPTIONS.csv
- DIAGNOSES_ICD.csv
- PROCEDURES_ICD.csv
- LABEVENTS.csv
- NOTEEVENTS.csv
- ADMISSIONS.csv
- There are six tables that need to be downloaded.:
- Download ndc2act, drug-atc,ndc2rxnorm_mapping data from the GAMENet
- torch >=1.7.1
- torch-cluster 1.5.8
- torch-geometric 1.0.3
- torch-scatter 2.0.5
- torch-sparse 0.6.8
- torch-spline-conv 1.2.1
- torch-tb-profiler 0.2.1
- torchaudio 0.7.2
- torchtext 0.2.1
- pytorch-pretrained-bert 0.4.0
- pytorch-transformers 1.2.0
- python >=3.6.2
We provide a simple demo for the medication recommendation task. Use the fine-tuned model saved_model. Run the demo_rec.py.
the script is:
python demo_rec.py \
--model_name fusion_ml \
--output_dir ./debug/ \
--graph \
--do_test \
--eval_batch_size 1 \
--data_name multilabel \
--predict_task rx \
--model_choice fusion_ml \
--saved_model_file $saved_model
There are the predicted result:
{
"id_1":{
"predication":["C01B","C03C","A12A","A12B","M01A","A06A","J01D","B05C","C02D","C01D","A03F","H04A","C01C","A01A","A10A","B01A","N07A","N02A","C07A","A12C","C10A","N01A","A02B","A02A","N02B"],
"targets":["C01B","C03C","A12A","A12B","M01A","A06A","J01D","B05C","C01D","A03F","H04A","C01C","A01A","A10A","B01A","N07A","C07A","A12C","A04A","C10A","N01A","C09C","A02B","A02A","N02B"
]
},
"id_2":{
"predication":["A06A","B05C","V03A","H04A","C08C","A01A","A10A","B01A","C07A","A12C","C10A","A02B","N02B"],
"targets":["B05C","V03A","H04A","C08C","A01A","A10A","B01A","C07A","A12C","C10A","A02B","A02A","N02B"
]
},
"id_3":{
"predication":["C03C","A12A","A12B","A06A","B05C","C01D","A03F","H04A","C01C","A01A","A10A","B01A","N07A","N02A","C07A","A12C","C10A","N01A","A02B","A02A","N02B"],
"targets":["C03C","A12A","A12B","A06A","J01D","B05C","C01D","A03F","C01C","A01A","A10A","B01A","N07A","N02A","C07A","A12C","C10A","N01A","A02B","N02B"
]
}
}
The pre-trained MedM-PLM saved in the MedM-PLM, the vocabulary of unstructured data at Text vocabulary and the vocabulary of structured data at Diagnosis code vocabulary and Medication code vocabulary.
code_tokenizer = EHRTokenizer('structured data dir')
txt_tokenizer = BertTokenizer.from_pretrained('unstructured data dir')
model = MedM_PLM.from_pretrain('MedM-PLM_dir', dx_voc=code_tokenizer.dx_voc,rx_voc=code_tokenizer.rx_voc)
Please contact liusicen@stu.hit.edu.cn for help or submit an issue.
@article{liu2022two,
title={Multimodal data matters: language model pre-training over structured and unstructured electronic health records},
author={Liu, Sicen and Wang, Xiaolong and Hou, Yongshuai and Li, Ge and Wang, Hui and Xu, Hui and Xiang, Yang and Tang, Buzhou},
journal={arXiv preprint arXiv:2201.10113},
year={2022}
}