The PNNL Proteomics Pipeline: P3
This project provides the basic analysis and tools for carrying out end-to-end proteomic analysis for data generated at PNNL. This repository will serve as a resource upon which we can build additional tools in a reusable fashion.
Overview
To contribute to this pipeline
Check out the CONTRIBUTING.md document. We welcome all additional tool and methods. Then you can create a pull request and merge your code in.
To use this pipeline
Click on the Use this template button above. This will allow you to create your own repository.
Pipeline overview
This pipeline links together a series of data and code repositories to ensure that any data is saved for future use. Furthermore we hope to be able to reproduce the analysis for future projects. A brief overview of each tool is described here.
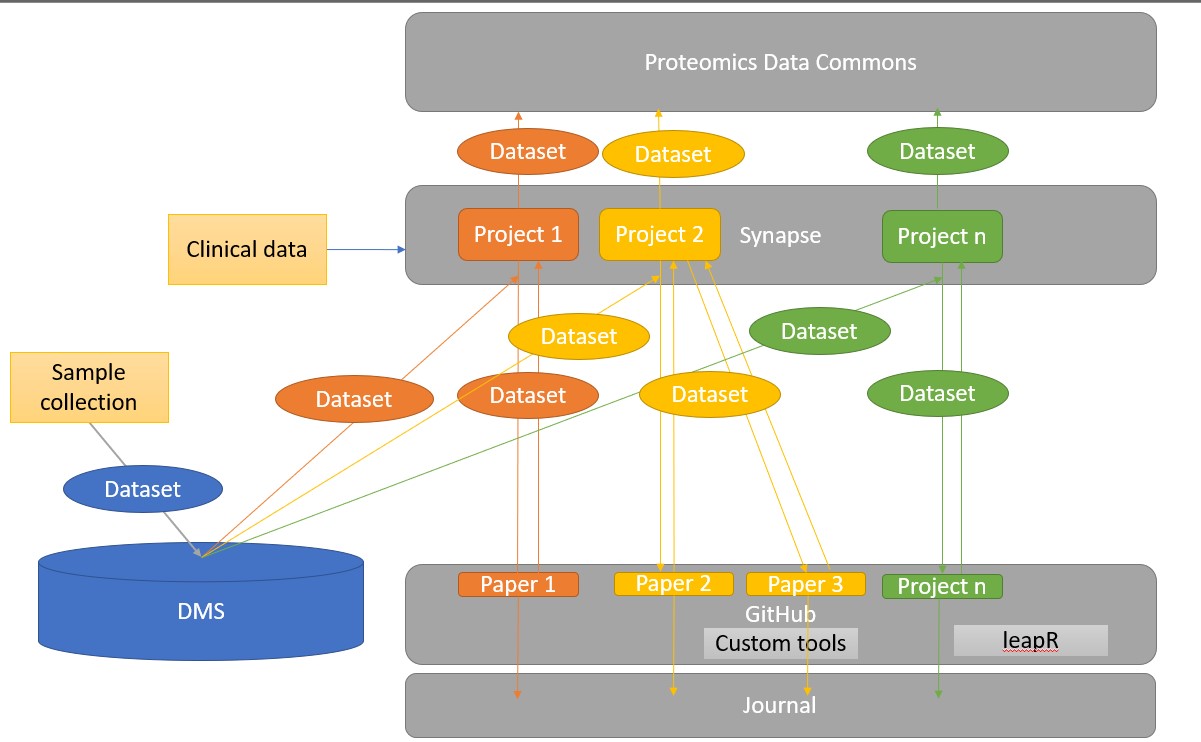
Proteomics and phosphoproteomics processing
This processing relies on a series of customized R scripts that are in the following directories:
- global: global proteomics analysis documentation is here
- phospo: phosphoproteomics analysis documentation is here Once this analysis is complete, the data should be stored on Synapse
Synapse processing
Synapse is used for:
- data storage: data is backed up for free
- data processing: UUIDs and permission modifications allow for easy analysis
- data release: data can be moved to the PDC or shared
You will need to acquire a synapse username to access synapse, and become a certified user to add data, but after that you will be set with future projects.
Functional analysis
There are typically a few steps we do for functional analysis. They are described in the functional analysis directory for now.