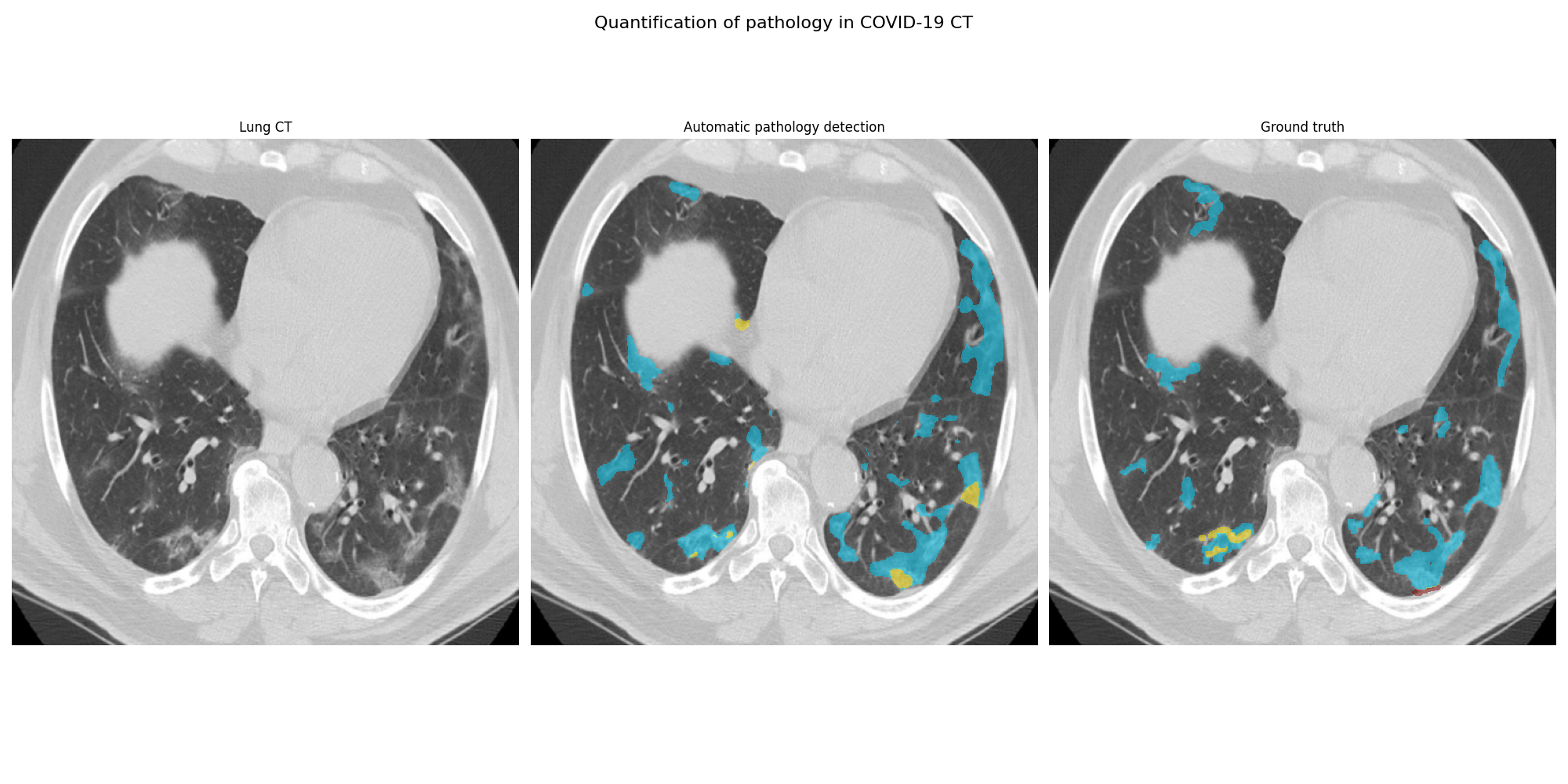
This repository provides proof of concept code for detecting pathology related to COVID-19 from a Sensyne hackathon. The figure below shows the original CT axial slice, the manually annotated ground truth and the automatic delineation of consolidation and ground glass opacity.
Please see the following blog post for more details: https://medium.com/@heaven_spring_cheetah_212/a-weekend-a-virtual-hackathon-and-ml-approaches-to-automate-the-analysis-of-covid-19-lung-ct-5ad4c89fe03c
This code is based on the U-Net implementation from: https://github.com/zhixuhao/unet
The original dataset is provided at:
https://www.sirm.org/2020/03/28/covid-19-caso-65/
The annotated dataset with annotations are available at from MedSeg
To run the code, download the training images, training masks and test images from the site (tr_im.nii.gz, tr_mask.nii.gz and val_im.nii.gz)
and copy these files into data/covid/ in the code directory.
The trained model can be downloaded from the the Github release at:
This needs to be copied into the main folder.
Create the virtual environment
python3 -m venv venv
pip install -r requirements.txt
To run the code
python3 main.py
docker build --tag covidruntime:1.0 .
To run the code
docker run --rm -it -v $(pwd):/code covidruntime:1.0 python3 main.py
The processed images from the test and validation set will appear in demo_figs
The following simple options are available in the main script:
# Options
# --------------------------------------
train_model = False
validation_size = 0.1
steps_per_epoch = 100
epochs = 200
# --------------------------------------Selecting train_model = True wil train the model from scratch.