This repository contains the training and inference code for my solution to the RSNA Abdominal Trauma Detection challenge, which placed 23rd on the public leaderboard and 24th on the private leaderboard. My solution consists of three stages:
- Organ segmentation
- Slice-level feature extraction
- Scan-level classification
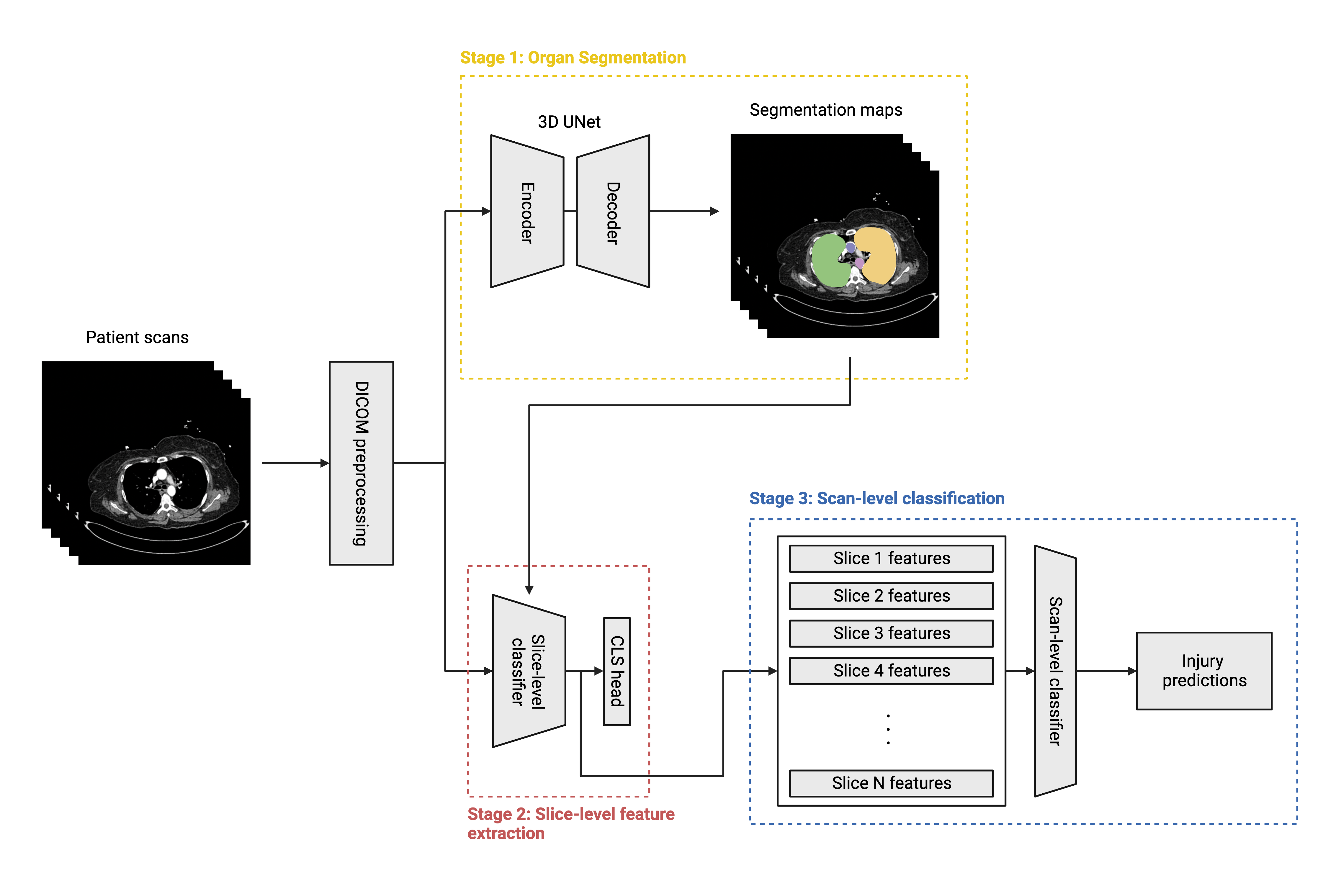
An overview of the solution is shown in the figure below. The full submission notebook is also included for reference.
The segmentation stage aims to extract bounds for relevant organs in the patient scans (bowel, liver, kidney, and spleen), which we can then leverage in downstream classification models. My choice of segmentator architecture is a 3D UNet model with an EfficientNet backbone and input dimensions of
# Training
python3 src/scripts/segmentation/train.py --config=configs/segmentation/tf_efficientnetv2_s_128_224_224.yaml
# Inference
python3 src/scripts/segmentation/train.py --config=configs/segmentation/inference.yaml
The second stage seeks to extract slice-level features from each patient scan, which are then fed to a scan-level classifier in the third stage. For this stage, I train segmentation-informed classification models on slice-level labels, which are derived by multiplying the scan-level labels by the organ visibilities for a given slice (for instance, a patient with a high-grade liver injury will have positive slice-level liver injury labels only on the slices where the liver is visible). I fine-tune three families of slice-level classifiers: EfficientNet, ConvNeXt, and MaxViT. During inference, I extract the
# Training
python3 src/scripts/slice_classification/train.py --config=configs/slice_classification/tf_efficientnetv2_s_384.yaml
# Inference
python3 src/scripts/slice_classification/infer.py --config=configs/slice_classification/inference.yaml
In the third and final stage, I concatenate the slice-level feature maps from the second stage into scan-level sequences. Thus, if a scan has
# Training
python3 src/scripts/slice_classification/train.py --config=configs/scan_classification/lstm_512_128.yaml
# Evaluation
python3 src/scripts/slice_classification/infer.py --config=configs/scan_classification/evaluation.yaml