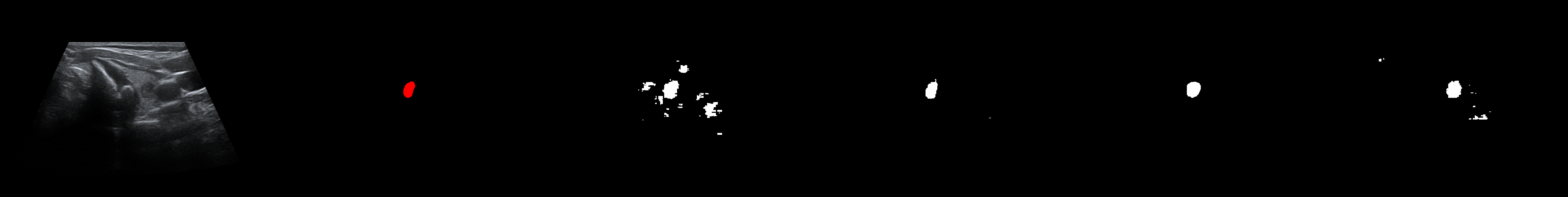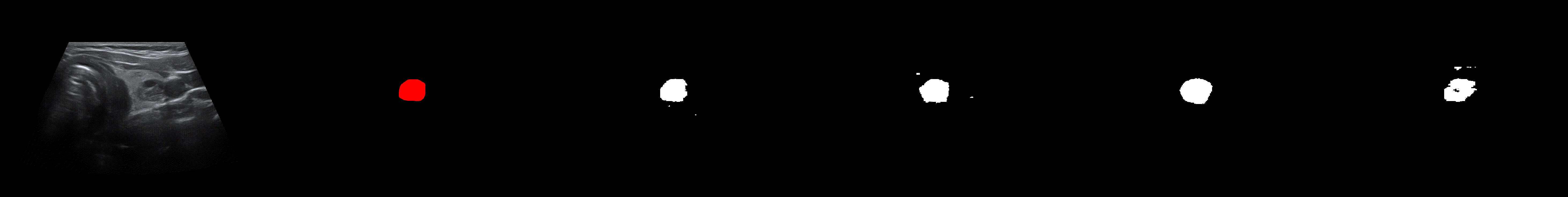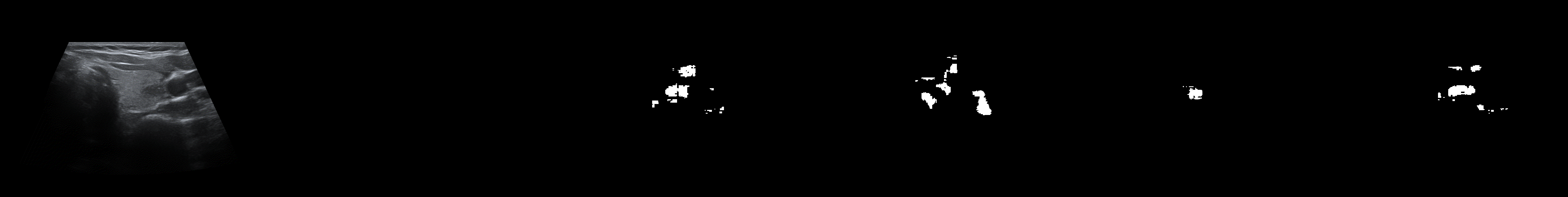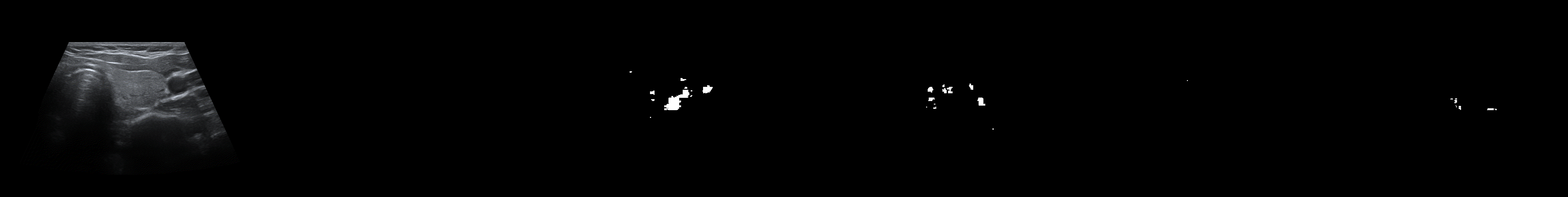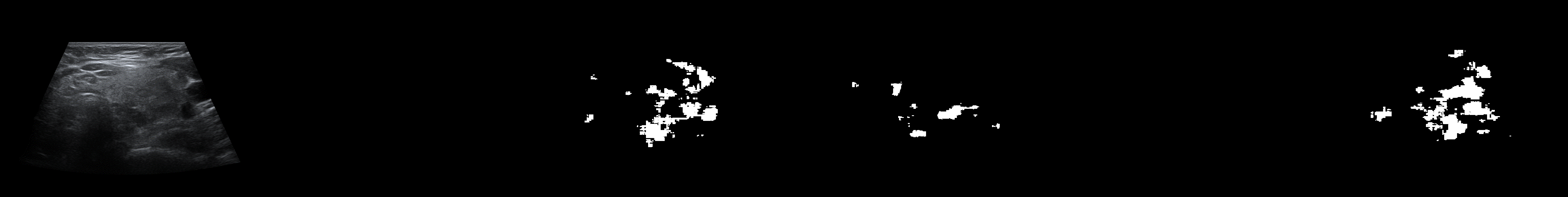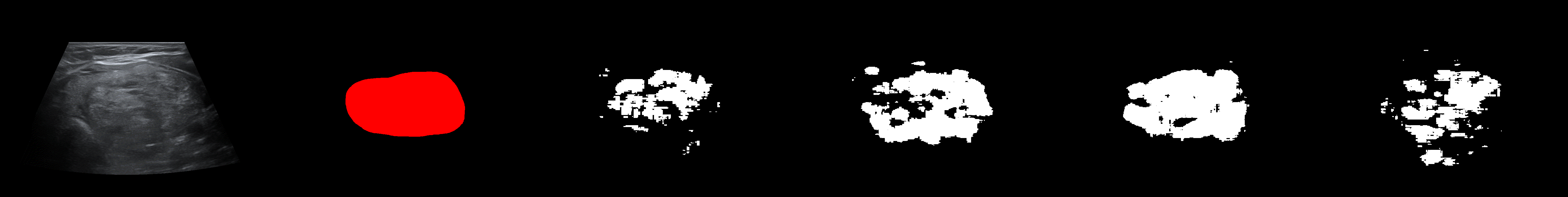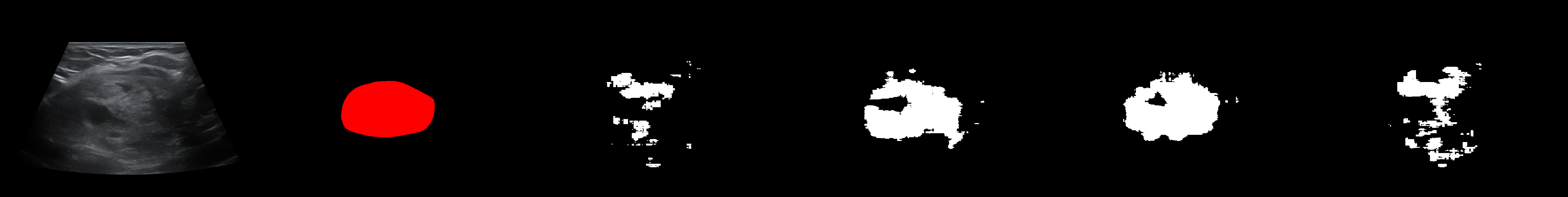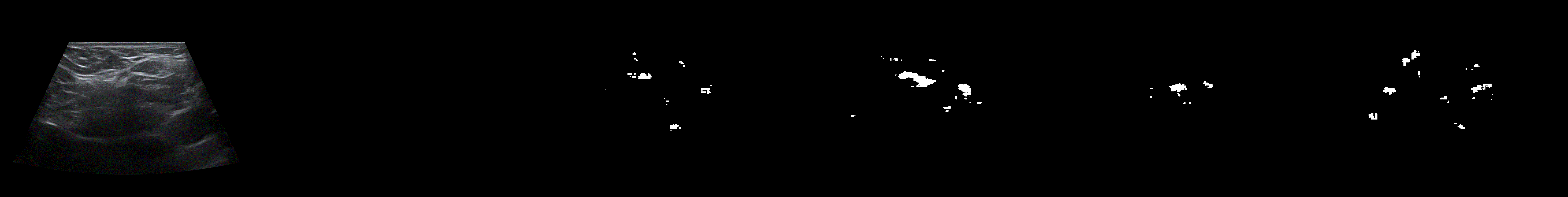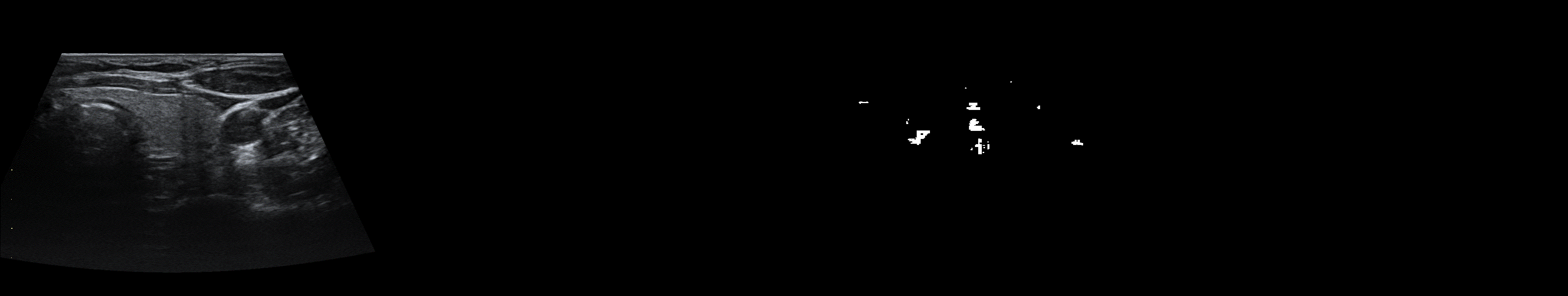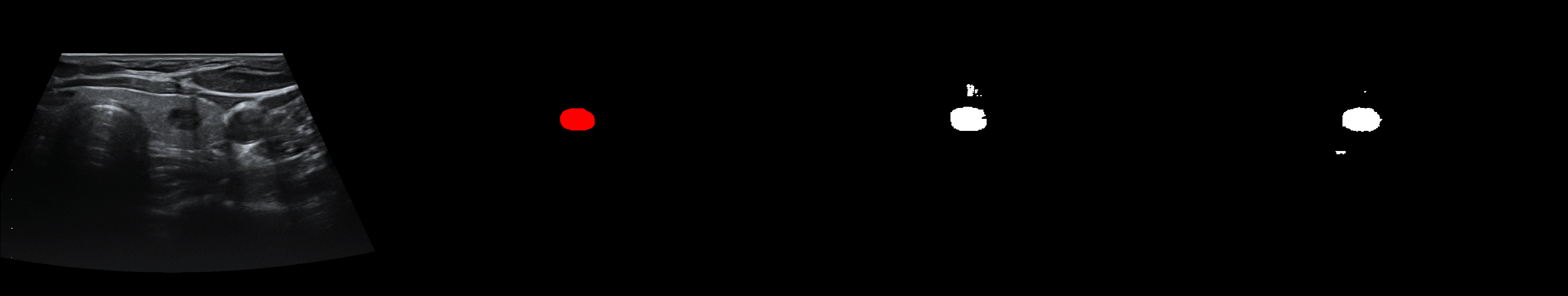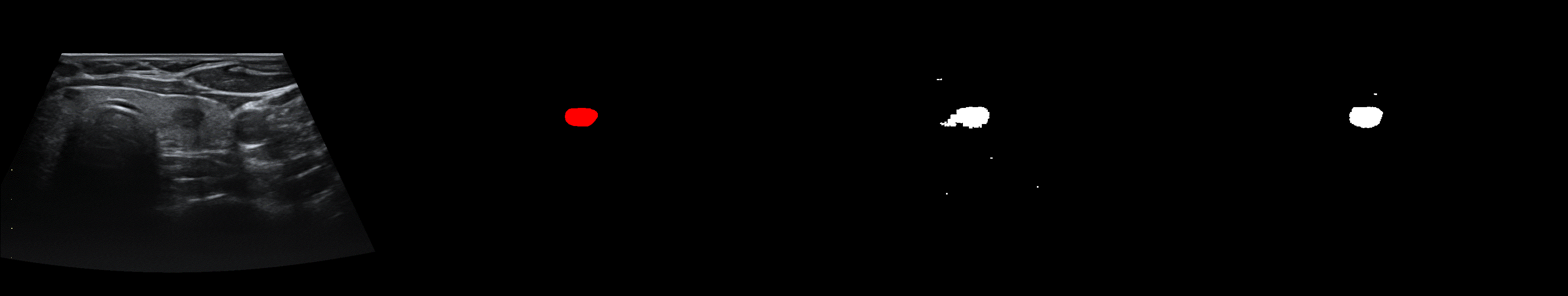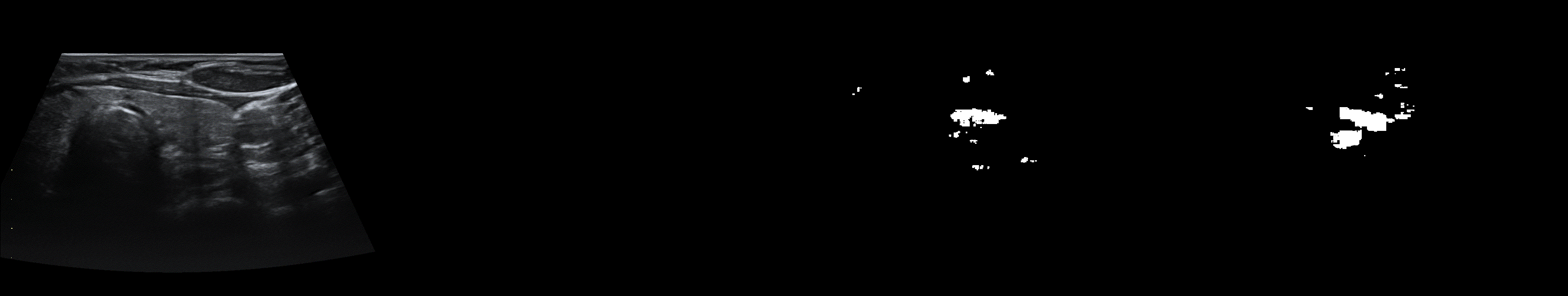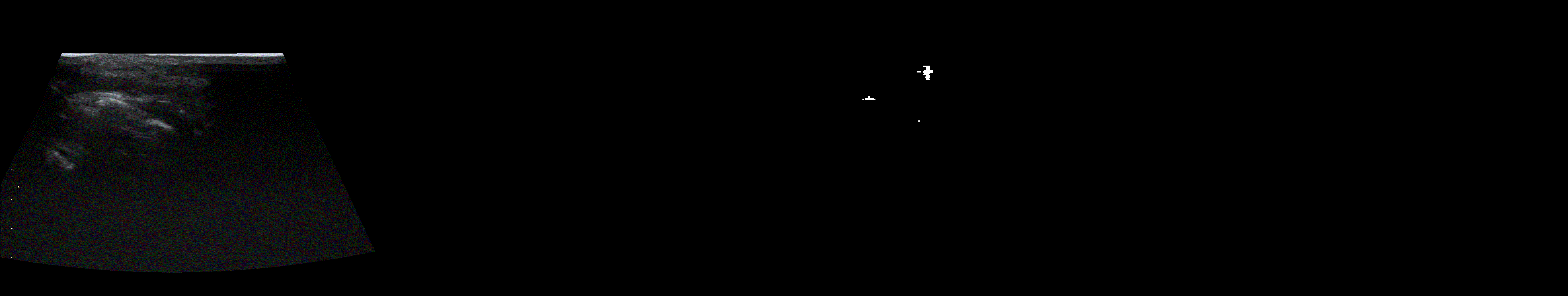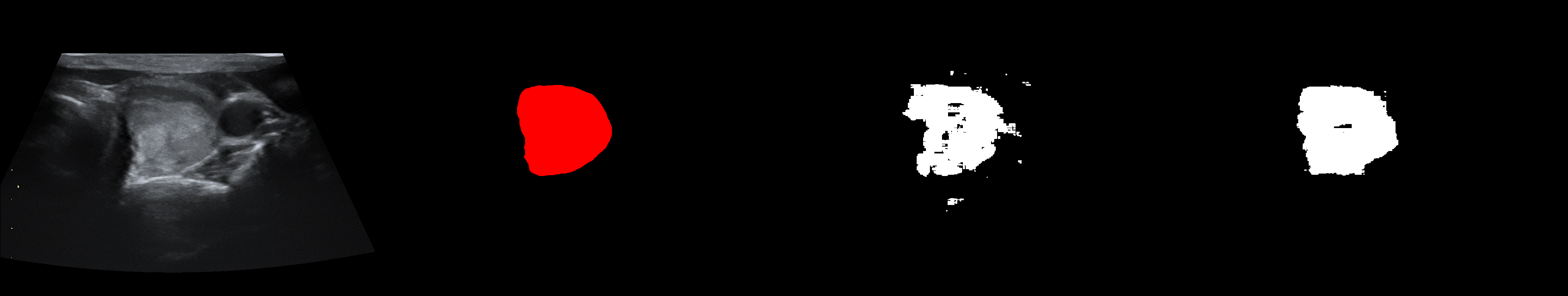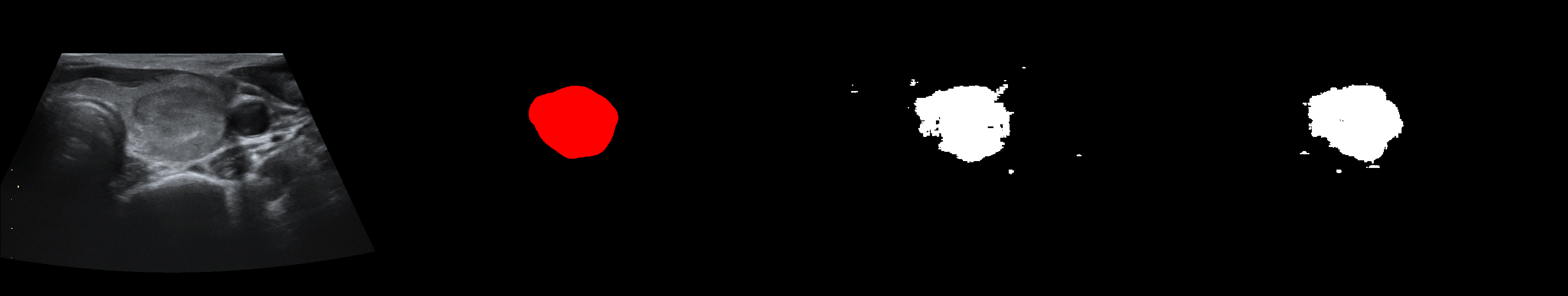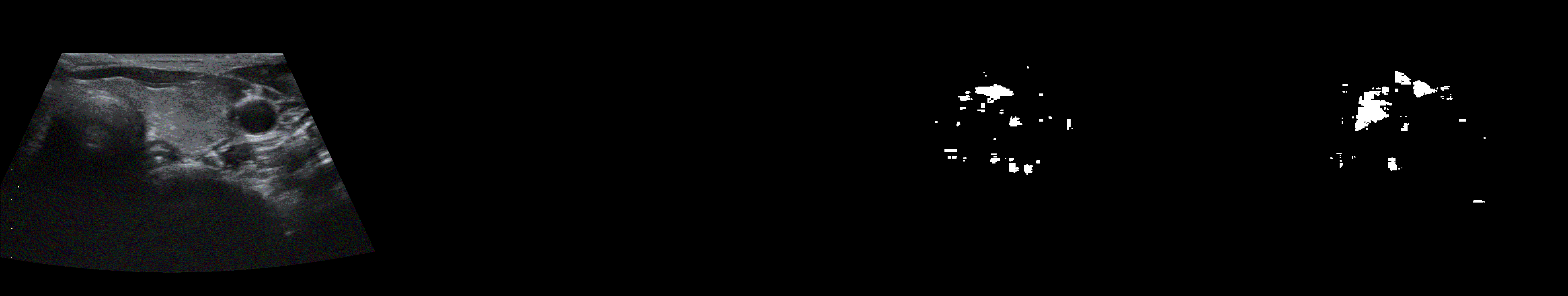This repository is dedicated to the implementation of the idea of the Global Adaptive Filtering Layer
Our contribution is that we try to explore the influence of inserting not one but several GAF layers into different parts of model. We experiment woth U-Net for segmentation and ResNet for classification and check metrics on BPUI and BUSI (open datasets with medical images)
- Nikita Kurduikov, @Pqlet
- Pavel TIkhomirov, @ocenandor
The file environment.yml contains the necessary libraries to run all scripts
It is necessary:
- create conda environment and install all packages through the command:
conda env create -f environment.yml - activate environment to run scripts from it:
conda activate ultrasound
To use a layer with your model, just do the following:
from models.adaptive_layer import AdaptiveLayer, GeneralAdaptiveLayer
class Model(torch.nn.Module):
def __init__(self, ..., n_channels=1, image_size=(512, 512), adaptive_layer_type=None):
super(UNet, self).__init__()
self.name = 'Model'
if adaptive_layer_type is not None:
self.name = '_'.join([self.name, 'adaptive', adaptive_layer_type])
self.adaptive_layer_type = adaptive_layer_type
if self.adaptive_layer_type in ('spectrum', 'spectrum_log', 'phase'):
self.adaptive_layer = AdaptiveLayer((n_channels, ) + image_size,
adjustment=self.adaptive_layer_type)
elif self.adaptive_layer_type == 'general_spectrum':
self.adaptive_layer = GeneralAdaptiveLayer((n_channels, ) + image_size,
adjustment=self.adaptive_layer_type,
activation_function_name='relu')
# your Model class init
def forward(self, x):
if self.adaptive_layer_type is not None:
x = self.adaptive_layer(x)
# your Model class forward method!Attention! The name of the folder with the dataset must be one of the following: Endocrinology, BUSI, BPUI
-
Endocrinology Dataset
This dataset provided by the Center for Endocrinology and is not in the public domain (there is no way to download it).
-
Breast Ultrasound Images Dataset (Dataset BUSI) (253 MB)
Al-Dhabyani W, Gomaa M, Khaled H, Fahmy A. Dataset of breast ultrasound images. Data in Brief. 2020 Feb;28:104863. DOI: 10.1016/j.dib.2019.104863.The archive should be unpacked and turned into the following structure:
├───BUSI ├───benign ├───benign (1).png ├───benign (1)_mask.png └─── ... ├───malignant ├───malignant (1).png ├───malignant (1)_mask.png └─── ... ├───normal ├───normal (1).png ├───normal (1)_mask.png └─── ... -
Brachial Plexus Ultrasound Images (BPUI) train (1.08 GB)
The archive should be unpacked and turned into the following structure:
├───BPUI ├───1_1.tif ├───1_1_mask.tif └─── ...
To train a model, you should
-
initially choose a CV task:
- Segmentation
- Classification
- Denoising/Erasing
-
correct
configs.yamlfile!Attention!Do not forget to specify path to folder with train data.!Attention!For classification task usenumber of classes = 3for BUSI dataset andnumber of classes = 6for Endocrinology dataset. -
for denoising/erasing task comment/uncomment lines in file
train_denoising.pywith appropriate noise overlaying transform -
run appropriate script
.py:
python ./train_<CV task>.py
or with parameters
- --device # cuda device id number (default 0)
- --random_seed # random seed (default 1)
- --nolog # turn off logging
As a result, you will receive
- the saved best model
.pthnext to files with code - a log file
.tfeventsof the training process in folder where the results of experiments from the tensorboard will be recorded (seeconfigs.yamlfile)
From left to right: ultrasound slice, ground truth mask, segmentation result performed by base model and with spectrum, spectrum log and phase adjustments.
From left to right: ultrasound slice, ground truth mask, segmentation result performed by base model and with general spectrum adjustment.
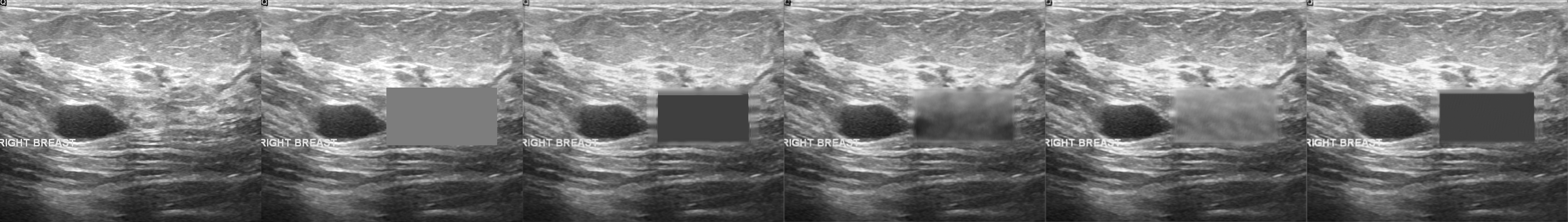
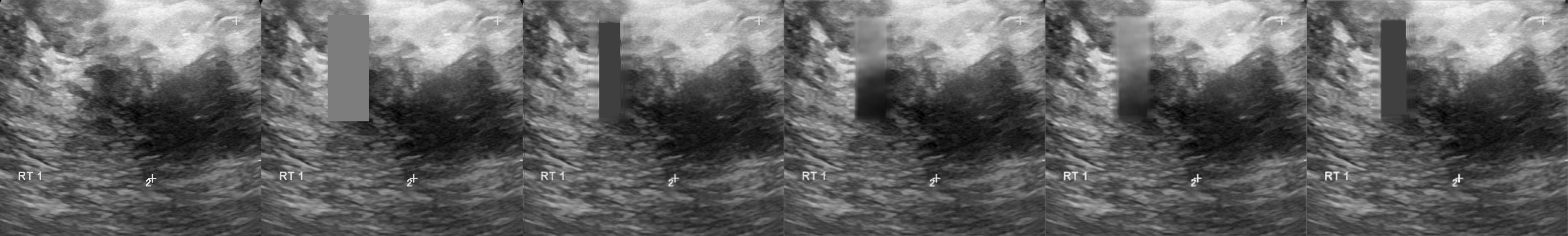
From left to right: ultrasound slice, corrupted image, restored image by base model and with spectrum, spectrum log and phase adjustments.
If you use this package in your publications or other work, please cite it as follows:
@article{2021_gafl,
author={Shipitsin, Viktor and Bespalov, Iaroslav and Dylov, Dmitry V},
year={2021},
month=Aug,
title={Global Adaptive Filtering Layer for Computer Vision},
journal={arXiv preprint arXiv:2010.01177v4},
}
Viktor Shipitsin - shipitsin@phystech.edu