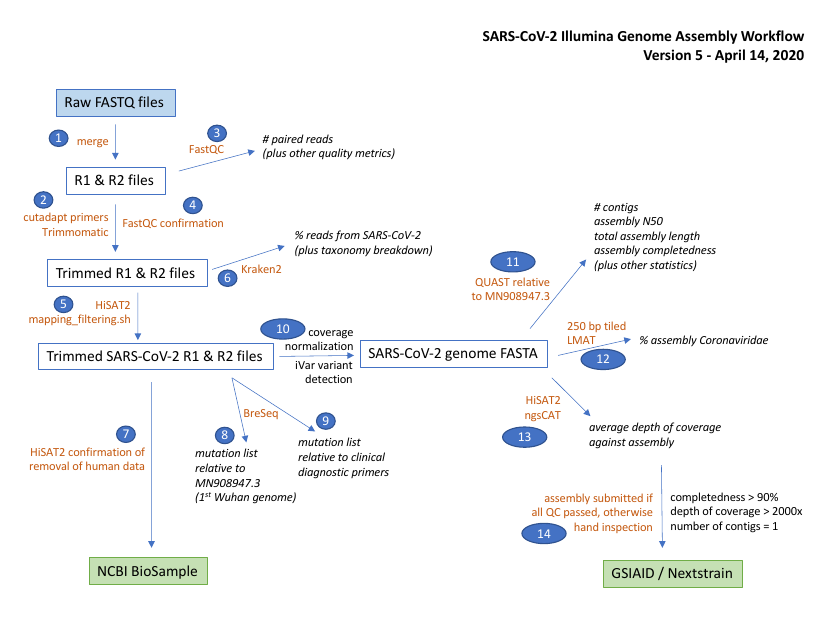
This snakemake pipeline is compatible with the illumina artic nf pipeline.
It performs the same consensus and variant calling procedure using ivar.
In addition it adds screening with Kraken2/LMAT, enhanced contamination removal, and additional breseq mutation detection.
By default the SARS-CoV2 reference genome: MN908947.3 is used throughout the analysis. This can be changed in the data dependencies download script (script/get_data_dependencies.sh) and updating the config.yaml accordingly. Similarly, the default sequencing primer and trimming settings can easily be added and adjusted in the config.yaml.
See below for full details.
Future enhancements are intended to aid/automate metadata management in accordance with PHA4GE guidelines, and manage upload to GISAID and INDSC compatible BioSamples.
-
Clone the git repository
git clone https://github.com/jaleezyy/covid-19-signal -
Install
condaandsnakemake(version >5) e.g.wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh bash Miniconda3-latest-Linux-x86_64.sh # follow instructions source $(conda info --base)/etc/profile.d/conda.sh conda create -n snakemake snakemake=5.11.2 conda activate snakemake
Additional software dependencies are managed directly by snakemake using conda environment files:
- fastqc 0.11.8 (docs)
- cutadapt 1.18 (docs
- trimmomatic 0.36 (docs)
- kraken2 2.0.7-beta (docs)
- unicycler 0.4.8 (github)
- quast 5.0.2 (docs)
- hisat 2.1.0 (docs)
- lmat, "sourceforge" version (sourceforge)
- samtools 1.7 (docs)
- bedtools 2.26.0 (docs)
- ivar 1.2 (docs)
- Download necessary database files
The pipeline requires:
- Amplicon primer scheme sequences (*)
- Nextera sequencing primer sequence files from trimmomatic
- SARS-CoV2 reference fasta
- SARS-CoV2 reference gbk
- SARS-CoV2 reference gff3
- kraken2 viral database
- LMAT
kML+Human.v4-14.20.g10.dbdatabase
All dependencies except the amplicon primers (*) can be automatically fetched using the follow accessory script:
bash pipeline/scripts/get_data_dependencies.sh -d data -a MN908947.3
- Configure your
config.yamlfile
Either using the convenience python script (pending) or
through modifying the pipeline/example_config.yaml to suit your system
- Specify your samples in CSV format (e.g.
sample_table.csv)
See the example table pipeline/example_sample_table.csv for an idea of how to organise this table.
-
Execute pipeline (optionally explicitly specify
--cores):snakemake --use-conda -s Snakefile --cores $(nproc) all
Alternatively, the pipeline can be deployed using Docker (see resources/Dockerfile_pipeline for specification).
To pull from dockerhub:
docker pull finlaymaguire/signal
Download data dependencies:
mkdir -p data && docker run -v $PWD/data:/data finlaymaguire/signal:1.0.0 bash scripts/get_data_dependencies.sh -d /data
Add remaining files (e.g. primers) to your config and sample table in the data directory:
cp config.yaml sample_table.csv $PWD/data && \
docker run -v $PWD/data:/data finlaymaguire/signal:1.0.0 mv data/config.yaml data/sample_table.csv .
Then execute the pipeline:
docker run -v $PWD/data:/data finlaymaguire/signal:1.0.0 conda run -n snakemake snakemake --use-conda --conda-prefix $HOME/.snakemake --cores 8 -s Snakefile all
- Generate summaries of BreSeq among many samples, see
For a step-by-step walkthrough of the pipeline, see pipeline/README.md.
A diagram of the workflow is shown below.