Question about reorientation
javak87 opened this issue · comments
Dear Developer,
I want to visually check around 100 CT images that are well aligned.
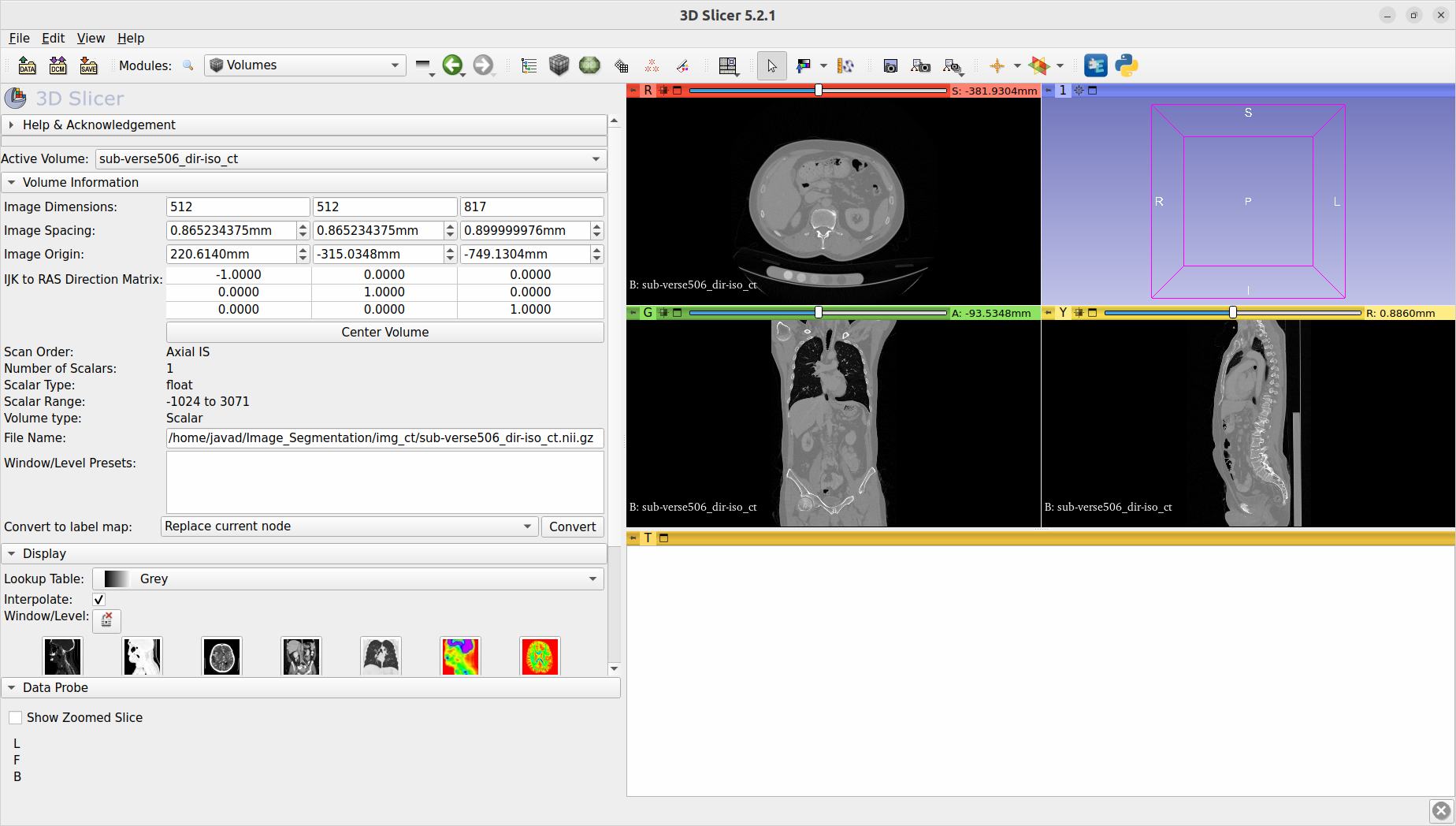
First, I pick one CT which looks good in the 3D slicer. I want to produce 3 views exactly like the 3d slicer view.
The coordinate system of the original CT is 'LAS'. I open this CT using 3d slicer and you can see the views in the following:
My task is to create views exactly like the above picture.
Since the 3D slicer works with 'RAS', First I convert to 'RAS' and create my images which are not simular to the 3D slicer.
This is my code:
scans_nib = nib.load('/home/javad/Image_Segmentation/img_ct/sub-verse506_dir-iso_ct.nii.gz')
orig_ornt = nib.io_orientation(scans_nib.affine)
targ_ornt = nib.orientations.axcodes2ornt("RAS")
transform = nib.orientations.ornt_transform(orig_ornt, targ_ornt)
img_orient = scans_nib.as_reoriented(transform)
scans_index = [img_orient.shape[0]//2, img_orient.shape[1]//2,img_orient.shape[2]//2]
img_0 = img_orient.get_fdata()[scans_index[0],:,:]
img_name_0 = 'img_00.jpg'
cv2.imwrite(img_name_0, img_0)
img_1 = img_orient.get_fdata()[:,scans_index[1],:]
img_name_1 = 'img_01.jpg'
cv2.imwrite(img_name_1, img_1)
img_2 = img_orient.get_fdata()[:,:,scans_index[2]]
img_name_2 = 'img_02.jpg'
cv2.imwrite(img_name_2, img_2)
img_0 = cv2.imread('img_00.jpg', cv2.IMREAD_GRAYSCALE)
img_1 = cv2.imread('img_01.jpg', cv2.IMREAD_GRAYSCALE)
img_2 = cv2.imread('img_02.jpg', cv2.IMREAD_GRAYSCALE)
According to this code, the result is:



As you can see, views related to my code and 3D slicer are different.
Could you please tell me how can I fix this problem?
Thanks
Since the 3D slicer works with 'RAS', First I convert to 'RAS' and create my images which are not simular to the 3D slicer.
I think this is a misunderstanding. Your images can be in any orientation and the affine shows how to convert the IJK indices into RAS coordinates. 3dSlicer will be using that to determine how to slice the image to find the view it wants. It is not simply showing you the middle slice of the first, second and third axes in RAS coordinates.
It would be good to start by figuring out what views you want. And it is important to note that numpy voxel arrays interpreted directly as images will proceed from top-to-bottom (x-axis), left-to-right (y-axis).
The first 3dSlicer image shows an slice that is ventral-dorsal in the x-axis and right-left in the y-axis.
I'm determining RL because your third image that is in RAS shows that the dots go from dark to light as your proceed left-to-right. Therefore I will assume all of these 3dSlicer views are radiological, not neurological.
The second image thus shows superior-inferior in the x-axis and right-left in the y-axis. And the third image clearly shows superior-inferior, ventral-dorsal.
Put together, if you want to directly slice your images in this order with the middle slice of the first axis oriented PL, the second IL and the third IP, then the orientation you need is IPL, not RAS.
Dear @effigies,
Great recommendation. I changed to IPL and it works.
My final task is, I have several CTs when I loaded in the 3D slicer, the sagittal, axial, and coronal are mixed.
I have to visually look at these images and changed the orientation such that the sagittal, axial, and coronal are ok in 3D scilcer.
Could you please tell me how can I fix this problem?
Thanks
I don't really understand the situation. Do the images have bad affines, or does 3D Slicer not respect the affine?
As I understood, images have bad affines because when I changed the other coordinates system like RAS or others, the 3D slicer automatically detect the views as shown before.
Thanks
If the images have bad affines, in the sense that the direction that is supposed to point left is not pointing left, then your only options are manual correction or finding the original data and fixing the conversion process to ensure that the affine is correctly set in the converted image.
If you just mean that Slicer 3D expects images to be oriented RAS and will show things in whatever orientation you get, then you can just use the process above to convert to whatever orientation is convenient for you to view it.
Thank you so much for your information.
If we have a bad affine and we don't have the original data, How can we make the affine correct manually?
I mean can I find the correct affine just by looking at scans_nib.header (image meta data) information?
can I find the correct affine just by looking at
scans_nib.header(image meta data) information?
Generally not. That is the source of the affine, and you said it's not correct.