Tired of writing the training code over and over, maintaining inconsistent versions of your code and model checkpoints or you just to want to write code that can be easily read used by other researchers?
Then what you need is Just Another DEep learning (JADE) framework.
JADE is a simple framework based on Weights and Biases and the PyTorch libraries designed to:
- Clearly separate the Model loss and logic from the Architectures, Dataset, Training procedure and Evaluation metrics, improving readability and re-usability of the code.
- Easily specify and tune hyper-parameters from configuration files
- Facilitate deployment on different (distributed) hardware. A deployment guide for clusters with SLURM scheduling can be found in the example.
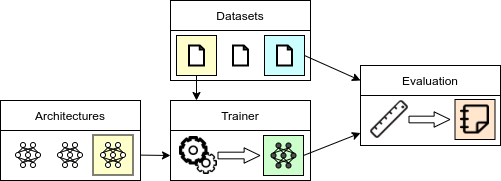
Each JADE project consists the specification of 4 main components:
- data: definition of the datasets used for training and evaluation.
- architectures: definition of the neural network architectures used by the model
- models: definition of model(s) and respective loss function(s). Each model as access and can instantiate the different architectures.
- trainers: definition of the training loop. Each trainer has access to the specified data and model.
- evaluators: definition of the evaluation metrics. Each evaluation metric has access to the model and data
The dependency between the various components is summarized in the following picture:
JADE can be installed using pip:
pip install jadeRun the initialization command
wandb initand enter your username and password as required.
The implementation of a new model consists of 3 steps, which are discussed in detail in the following sections:
├── architectures
│ ├── MNIST.py
│ └── CIFAR10.py
├── datasets
│ ├── MNIST.py
│ └── CIFAR10.py
├── evaluation
│ ├── accuracy.py
│ └── ELBO.py
├── models
│ └── VAE.py
└── trainers
- Definition the model. The implementation must extend the
jade.Modelclass. More details can be found in the section Model definition. Each model definition must be included in amodels/folder. Note that each model should be designed to be independent of architectures and data.
import jade
class MyModel(jade.Model):
def initialize(self, **model_and_arch_params):
# Initialization of the architectures and hyper-parameters
def compute_loss(self, data_batch):
# Loss computation- Definition of the required (neural networks) architectures (section Architectures).
Architecures must be included in a
/architecuresfolder. Different implementation could correspond to different modeling strategies or different datasets.
import torch.nn as nn
class MyArchitecture(nn.Module):
def __init__(self, **arch_params):
# Model code here- Definition the datasets (only if not already supported by PyTorch, section Dataset. Each definition needs
to be included in a
datasets/folder.
from torch.utils.data import Dataset
class MyDataset(Dataset):
def __init__(self, **dataset_params):
# Dataset code here- Definition of the evaluation metrics. Each metric must extend the
jade.Evaluationclass (section Evaluation)). The output ov the evaluation metrics is logged directly on wandb. Evaluation metrics must be included in the\evaluationfolder.
import jade
class MyEvaluation(jade.Evaluation):
def initialize(self, **eval_params):
# Initialization of the evaluation metrics
def evaluate(self):
# Evaluation code here- Write the model configuration, dataset and evaluation .yaml file to specify the respective hyperparameters. (section
Parameters confituration) - Run the model or define a sweep file for testing hyper-parameter ranges (section
Running the experiments).
Each new Model and algorithm must extend the core.Trainer class and define the architecture initialization and training.
The models need to override the default initialize(**params) and train_step(data) as discussed in the following sections.
The framework is designed to make sure each model implementation is agnostic of the dataset and architectures to separate
the logic of the algorithm from its implementation details.
Place the implementations of your models in the modules/models folder.
All arguments of the initialize(**params) method can be directly specified in the corresponding configuration file (see section Parameters confituration/Model and architectures),
therefore all the parameters that needs to be explored can be added to the method signature.
# Example of the Variational Autoencoder Model https://arxiv.org/abs/1312.6114
# Content of the models/VAE.py file
class VariationalAutoencoder(Trainer):
def initialize(self, z_dim, encoder_layers, decoder_layers, beta, lr, batch_size, n_workers=0, sigma=1):
# The value of the z_dim, n_encoder_layers, n_decoder_layers, beta lr, batch_size and n_workers are defined
# in the configuration file or specified as additional arguments when running the train python file
# Store the value of the regularization strength beta for the loss computation
self.beta = betaWithin the initialize method, the self.instantiate_architecture(class, params) method can be called to instantiate
the corresponding class defined in the Architecture file (see section Architectures) with the specified parameters.
# initialize the encoder "Encoder(z_dim, layers)" defined in the architecture python file
self.encoder = self.instantiate_architecture('Encoder',
z_dim=z_dim, layers=encoder_layers)
# initialize the encoder "Decoder(z_dim, layers, sigma)" defined in the architecture python file
self.decoder = self.instantiate_architecture('Decoder',
z_dim=z_dim, layers=decoder_layers, sigma=sigma)
# Initialize the Gaussian prior passing the number of dimensions
self.prior = self.instantiate_architecture('Prior',
z_dim=z_dim)
# Initialize the optimizer passing the parameters of encoder, decoder and the specified learning rate
self.opt = Adam([
{'params': self.encoder.parameters(), 'lr':lr},
{'params': self.decoder.parameters(), 'lr':lr},
])All the attributes (and architectures) that needs to be stored/restored can be specified by calling the
self.add_attribute_to_store(attribute_name) method
# Store the parameters of encoder, decoder and optimizer
self.add_attribute_to_store('encoder')
self.add_attribute_to_store('decoder')
self.add_attribute_to_store('opt')The attribute self.train_loader also needs to be instantiated within the initialize method to specify the data loader
used during training. The datasets defined in the Dataset configuration files are available by accessing the
self.datasets[dataset_name] attribute (see section Parameters confituration/Dataset)
# Instantiate a default pytorch DataLoader using the 'train' dataset defined in the dataset confituration file
self.train_loader = DataLoader(self.datasets['train'],
batch_size=batch_size, n_workers=n_workers, shuffle=True)Each model needs to define a train step that will receive a batch of data from the train loader for each new iteration.
def train_step(self, data):
# Note that the data is already moved to the device of the trainer and all the architectures are set to train mode
x = data['x']
rec_loss, reg_loss = self.compute_loss_components(x)
# Add the two loss components to the log
# Note that only scalars are currently supported although custom logging can be implemented
# by extending the implemented methods
self.add_loss_item('Rec Loss', rec_loss.item())
self.add_loss_item('Reg Loss', reg_loss.item())
loss = rec_loss + self.beta * reg_loss
self.opt.zero_grad()
loss.backward()
self.opt.step()
def compute_loss_components(self, x):
# Encode a batch of data
q_z_given_x = self.encoder(x)
# Sample the representation using the re-parametrization trick
z = q_z_given_x.rsample()
# Compute the reconstruction distribution
p_x_given_z = self.decoder(z)
# The reconstruction loss is the expected negative log-likelihood of the input
# - E[log p(X=x|Z=z)]
rec_loss = - p_x_given_z.log_prob(x).mean()
# The regularization loss is the KL-divergence between posterior and prior
# KL(q(Z|X=x)||p(Z)) = E[log q(Z=z|X=x) - log p(Z=z)]
reg_loss = (q_z_given_x.log_prob(z) - self.prior().log_prob(z)).mean()
return rec_loss, reg_lossDifferent quantities can be easily added to the log by calling the self.add_loss_item(name, value).
The values will be added to the corresponding tensorboard log. The logging frequency can be reduce by setting the
log_loss_every variable (see section Running the experiments).
# Add the two loss components to the log
# Note that only scalars are currently supported although custom logging can be implemented
# by extending the implemented methods
self.add_loss_item('Rec Loss', rec_loss.item())
self.add_loss_item('Reg Loss', reg_loss.item())The following methods can be extended if required:
on_start(): method called only once after initializationon_iteration_end(): method called at the end of each training step. Don't forget to increment the iteration count if this is overwrittenon_epoch_end(): method called once at the end of each training epoch. Don't forget to increment the epoch count if this is overwritten
Each trainer also has a end_training() method that can be called within the training procedure or by any evaluator to
terminate the training procedure.
The different architectures that are instantiated in the Trainer described in the previous section needs to be specified in the 'architectures_file'.
# Content of the architectures/simple_MNIST.py file
# Model for p(Z|X)
class Encoder(nn.Module):
def __init__(self, z_dim, layers):
super(Encoder, self).__init__()
# Create a stack of layers with ReLU activations as specified
nn_layers = make_stack([N_INPUT] + layers)
self.net = nn.Sequential(
Flatten(), # Layer to flatten the input
*nn_layers, # The previously created stack
nn.ReLU(True), # A ReLU activation
StochasticLinear(n_hidden, z_dim, 'Normal') # A layer that returns a factorized Normal distribution
)
def forward(self, x):
# Note that the encoder returns a factorized normal distribution and not a vector
return self.net(x)
# ... The definition of Decoder are analogous and can be found in the `architectures/simple_MNIST.py` fileNote that the parameters can be passed when calling the instantiate_architecture in the model definition.
IMPORTANT: while models and datasets are automatically imported by the framework, the architecture file are passed when
running the training scipt. This allows the same architecture (e.g. Encoder) to have completely different implementations
for different datasets.
The default torchvision datasets are supported by the framework. We recommend to create a simple Wrapper to make sure the Dataset object accepts only simple arguments such as float, strings, list and dictionaries as parameters so that the dataset can be described with a simple yaml configuration file.
# Content of examples/example/data/MNIST.py
MNIST_TRAIN_EXAMPLES = 50000 # Size of the training set (the rest is used for validation)
class MNISTWrapper(Dataset):
def __init__(self, root, split, download=False):
# We add an extra argument 'split' to the signature to create a the validation set
assert split in ['train', 'valid', 'train+valid', 'test']
# Add the ToTensor() transform to convert PIL images into torch tensors
dataset = MNIST(
root=root,
train=split in ['train', 'valid', 'train+valid'],
transform=ToTensor(), download=download)
if split == 'train':
dataset = Subset(dataset, range(MNIST_TRAIN_EXAMPLES))
elif split == 'valid':
dataset = Subset(dataset, range(MNIST_TRAIN_EXAMPLES, len(dataset)))
elif not (split == 'test') and not (split == 'train+valid'):
raise Exception('The possible splits are "train", "valid", "train+valid", "test"')
self.dataset = dataset
def __getitem__(self, item):
x, y = self.dataset[item]
return {'x': x, 'y': y}Unless a wrapper is required as in the example reported above to conver PIL images to tensors, torchvision classes can be referred directly from the configuration file.
The Jade framework allows for the definition of arbitrarily evaluators used for logging purposes.
Each evaluator must extend the jade.eval.Evaluaiton class and extend the initialize(**params) and evaluate() methods.
Each evaluator has access to all the dataset definitions and the trainer object that can be easily accessed as attributes.
The initialize(**params) method is used for the initialization of the evaluator and receives the parameters form the
evaluation configuration file.
# Code from example/eval/elbo.py
class ELBOEvaluation(Evaluation):
def initialize(self,
evaluate_on, # name of the dataset on which the metric is evaluated
n_samples=2048, # number of evaluations to compute the expectation
resample_x=False, # use the same x over time if false, otherwise shuffle randomly
batch_size=256 # batch size used for evaluation
):
# Make a data_loader for the specified dataset (names are defined in the dataset configuration file).
# with a default batch size of 256
self.data_loader = DataLoader(self.datasets[evaluate_on], shuffle=resample_x, batch_size=batch_size)
self.n_samples = n_samples
self.batch_size = batch_size
# Check that the model has a definition of the function used to compute training and evaluation errors
if not hasattr(self.trainer, 'compute_loss_components'):
raise Exception('The trainer must have a function `compute_loss_components` that returns reconstruction and regularization errror')
def evaluate(self):
evaluations = 0.
elbo = 0.
# Consider the device on which the model is located
device = self.trainer.get_device()
# Enable evaluation mode
self.trainer.eval()
# Disable gradient computation
with torch.no_grad():
for data in self.data_loader:
# Move the data to the corresponding device
x = data['x'].to(device)
# Compute the two loss components
rec_loss, reg_loss = self.trainer.compute_loss_components(x)
elbo += rec_loss.sum() + reg_loss.sum()
evaluations += self.batch_size
if evaluations >= self.n_samples:
break
return {
'type': 'scalar', # Type of the logged object, to be interpreted by the logger
'value': elbo/evaluations, # expectation to log
'iteration': self.trainer.iterations # Iteration count at the point of logging
}
# The definition of the ImageReconstructionEvaluation class in analogous and reported in the example/eval/rec_error.py file
# ...The frequency at which each evaluation is produced can be also specified from the evaluation configuration file. At the moment only support for scalars and figures has been added, but the interface can be easily adapted to fit any other data-type which is supported by tensorboard/wandb.
The value of all the parameters for model training, dataset specification and evaluation needs to be included into 3
respective .yml configuration files. Note that in each .yaml configuration file it is possible to refer to enviroment
variables with the syntax $VARIABLE_NAME (or ${VARIABLE_NAME}). This allows to easily specify the location of datasets and devices to use
on different machines.
The dataset configuration file contains the description of the class and parameters for the definition of the datasets
that are used during training and evaluation (validation and test).
The file describes a dictionary in which each key represent a different dataset (e.g. train and test) with the
respective parameters.
We define a configuration for the dataset used during validation and hyper-parameters search
# Content of configurations/data/MNIST_valid.yml
# Define a dataset that will be identified with the key 'train'
train:
# Class that will be instantiated (see the MNIST wrapper defined in the section above)
class: MNISTWrapper
# List of parameters that will be passed to the MNISTWrapper constructor
params:
# Note that the value of the environment variable DATA_ROOT will be replacing the corresponding token
root: $DATA_ROOT
split: train
download: True
# We test a disjoint validation set which will be refered to as 'test'
test:
class: MNISTWrapper
params:
root: $DATA_ROOT
split: valid
download: True
# Other splits or datasets can be defined here with arbitrary keys.Analogously, we can define a configuration for the final training (after the hyper-parameter search) in which the model is trained on train+validation set and tested on the test set
# Content of configurations/data/MNIST_final.yml
train:
class: MNISTWrapper
params:
root: $DATA_ROOT/datasets/MNIST
split: train+valid # Train the model on validation + train set
download: True
test:
class: MNISTWrapper
params:
root: $DATA_ROOT/datasets/MNIST
split: test # Test on the test set
download: True At training time we can easily specify which one of the two configurations is used by specifying the respective
configuration file (see section Running a model).
Note that datasets that are not referred during training or evaluation will not be instantiated to avoid
unnecessary memory consumption, while datasets that are referred (with self.datasets[<key>] in the Trainer or Evaluation
modules) need to be described in the configuration file.
Analogously to the dataset file, the evaluation description .yml file contains a dictionary of evaluation metrics that
are considered during training.
Each evaluator will have the evaluate_every parameters other than the ones defined in the initialize(**params) method,
which specifies the frequency (in epochs) at which the evaluation function will be called.
# Configuration in configurations/eval/VAE_simple.yml
# The key will be used as a name in the tensorboard/wandb log
TrainReconstructionsError:
class: ELBOEvaluation # We log the expected negative variational lower bound
# Parameters of the initialize(**params) method of the ReconstructionEvaluation class
params:
evaluate_on: train # On the training set
# Analogously, we define an evaluator for the ELBO of 'test' pictures
# (coming either from the validation or test set depending on the specified dataset configuration)
TestReconstructionsError:
class: ELBOEvaluation # The same metric is computed on the test dataset
params:
evaluate_on: test
# We also log the image reconstruction on train and test dataset using the ReconstructionEvaluation
# defined in example/eval/image_eval.py
TrainImageReconstructions:
class: ReconstructionEvaluation
params:
evaluate_on: train
n_pictures: 10
sample_images: False
# Frequency of evaluation (in epochs) if not specified, the default value is 1
evaluate_every: 2
TestImageReconstructions:
class: ReconstructionEvaluation
params:
evaluate_on: test
n_pictures: 10
sample_images: False
evaluate_every: 2Analogously we can write an evaluation description file for the final evaluation (model trained on train+evaluation and tested on test) just by defining a second evaluation file (see `configurations/eval/VAE_simple_final.yml)
Each model congiguration file includes a class descriptor that refers to a Trainer definition
and the description of its parameters:
# Configuration in configurations/models/VAE.yml
class: VariationalAutoencoderTrainer
params:
#################
# Architectures #
#################
# Encoder
encoder_layers:
-1024
-256
# Decoder
decoder_layers:
- 256
- 1024
###########################
# Optimization parameters #
###########################
# Learning Rate
lr: 0.0001
# Size of the training batch
batch_size: 256
# Read the number of workers from the environment variable
num_workers: ${NUM_WORKERS}
##########################
# Other model parameters #
##########################
# regularization strength
beta: 0.5
# Number of latent dimensions
z_dim: 64In order to run a model launch the script train.py specifying the following flags
--data_conf: path to the data configuration.ymlfile--eval_conf: path to the data configuration.ymlfile--model_conf: path to the model configuration.ymlfile--arch_impl: path to the architecture implementation.pyfile
Additional flags can be used to specify the device on which to run the model, maximum number of training epochs,
random seeds and model checkpoint frequency. For more information use the --help flag.
The flags '--device' and '--experiments-root' referring to the device on which the model is trained and the directory
in with the trained models and backups are stored can be also specified by setting the environment variables DEVICE
and EXPERIMENTS_ROOT respectively.
The whole JADE framework has been designed to facilitate parameters sweeps using the wandb library. When writing a wandb sweep file, the parameters in the model, data and evaluation configurations can be easily accessed:
# File in sweeps/simple_vae_sweep.yml
# Example of a sweep for the beta parameter and latent size of the Variational Autoencoder model
command:
- ${env}
- ${interpreter}
- ${program}
# Flags for the train.py script as explained above
- --data_conf=configurations/data/MNIST_valid.yml
- --eval_conf=configurations/eval/VAE_simple.yml
- --model_conf=configurations/models/VAE.yml
- --arch_impl=example/architectures/simple_MNIST.py
- --epochs=10
- --seed=42
method: random
parameters:
# Model parameters can be accessed b${env}y using the operator '.' to access the members of dictionaries
# to access the parameters 'beta' in 'params' of a model (see the model configuration file) we use the syntax
# model.params.beta as shown below
model.params.beta:
# We sample the parameter beta between e^-10 and e^1 using a log-uniform distribution
distribution: log_uniform
min: -10
max: 1
# We can also specify a value (or list of values) directly. This is going to override the default specified
# in the model configuration file
model.z_dim:
values:
- 32
program: train.pyThe parameters defined in the model configuration file can be accessed using model.params.<PARAMETER_NAME>.
Analogously, the parameters regarding data and evaluation configuration can be accessed using params.<PARAMETER_NAME>
and eval.<EVAL_NAME>.params.<PARAMETER_NAME>respectively.
Another example of a sweep configuration file for the bayesian hyper-parameter search on a
Variational Information Bottleneck can be found in sweeps/simple_vib_sweep.yml.
Once the sweep .yml file has been defined, run the corresponding wandb command to create a sweep
wandb sweep <sweep_file>
The command will return a sweep_id that can be used to start the corresponding agents. Note that the sweep_id
wandg agent <wandb_user/wandb_project/sweep_id>
Note that conveniently the agents can be launched from different machines at the same time on the same sweep. For each machine running an agent make sure that:
- the code version and environment are consistent and updated
- wandb has been initialized with the
wandb initcommand - all the environment variables defined in the
.ymlfiles have been defined together withWAND_USERandWANDB_PROJECT
While the sweep declaration can be submitted from a local machine, running the agent usually requires a certain amount of coumputational resources.
Here we report the procedure to deploy and run the agents on a cluster using SLURM for scheduling. 0) install conda on your cluster node. This will make it easier to deal with dependencies
- make sure to download the code at the most recent version. If you are using git, you can simply run a
git cloneorgit pullto retrieve the most updated version of the code - install the conda environment
conda env create -f environment.yml - make sure wandb is accessible and initialized with
wandb init - create a .sbatch file for SLURM (see the documentation here ). here we report an example
#!/bin/bash
# SLURM flags (see https://slurm.schedmd.com/sbatch.html)
# Resource allocation
#SBATCH --gres=gpu:1
#SBATCH --mem=10G
#SBATCH --cpus-per-task=16
#SBATCH --time 10:00:00
#SBATCH --ntasks=1
# Other parameters
#SBATCH --priority=TOP
#SBATCH --job-name=<JOB_NAME>
#SBATCH -D <YOUR_HOME_DIRECTORY>
#SBATCH --output=log.txt
#SBATCH --verbose
# Cuda variables
export CUDA_HOME=/usr/local/cuda-10.0
export PATH=${CUDA_HOME}/bin:${PATH}
export LD_LIBRARY_PATH=${CUDA_HOME}/lib64:$LD_LIBRARY_PATH
export CUDA_CACHE_PATH="$TMPDIR"/.cuda_cache/
export CUDA_CACHE_DISABLE=0
# Setting the environment variables
# Weights and biases username and password
export WANDB_USER=<WANDB_USERNAME>
export WANDB_PROJECT=<WANDB_PROJECT>
# Parameters for number of cores for data-loading and device on which the model is placed
export NUM_WORKERS=16
export DEVICE=cuda
# Pointing to the directory in which the dataset is stored
export DATA_ROOT=<DATASET_ROOT> #(e.g. /hddstore/datasets)
# We use temp to save the backups since they are uploaded and deleted afterwards
mkdir /tmp/experiments
export EXPERIMENTS_ROOT=/tmp/experiments
#Generate cuda stats to check cuda is found
nvidia-smi
echo Starting
# Make sure you are in the project directory before trying to run the agent (the one containing the train.py file)
cd <PROJECT_DIRECTORY>
# Use python from the pytorch_and_friends environment
export PATH=$HOME/anaconda3/envs/pytorch_and_friends/bin/:$PATH
# Check that python and wandb are accessible
which python
which wandb
# Run the agent
echo Starting agent $WANDB_USER/$WANDB_PROJECT/$SWEEP_ID
wandb agent $SWEEP_ID
wait
# Remove all the files for the model backups since wandb is uploading them anyway
# You can change the experiments directory if you want to keep local versions
rm -r /tmp/experiments
- Submit the SLURM job
sbatch --export=SWEEP_ID=<YOUR_SWEEP_ID> <YOUR_SLURM_FILE.sh>
The console output and updated results for your agents can be found on the wandb website.