Omkar Thawakar* , Abdelrahman Shaker* , Sahal Shaji Mullappilly* , Hisham Cholakkal, Rao Muhammad Anwer, Salman Khan, Jorma Laaksonen, and Fahad Shahbaz Khan.
*Equal Contribution
Mohamed bin Zayed University of Artificial Intelligence, UAE
- Aug-04 : Our paper has been accepted at BIONLP-ACL 2024 🔥
- Jun-14 : Our technical report is released here. 🔥🔥
- May-25 : Our technical report will be released very soon. stay tuned!.
- May-19 : Our code, models, and pre-processed report summaries are released.
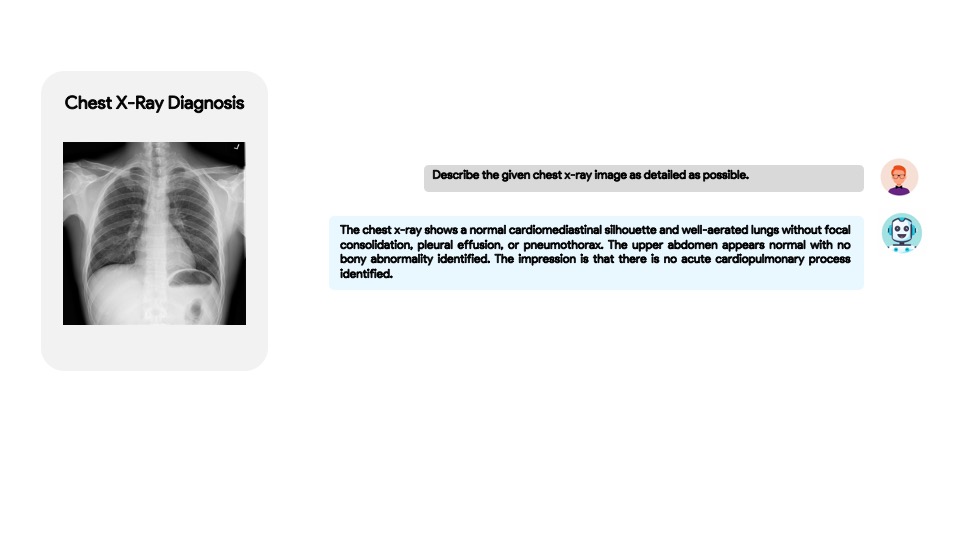
You can try our demo using the provided examples or by uploading your own X-ray here : Link-1 | Link-2 | Link-3 .
- XrayGPT aims to stimulate research around automated analysis of chest radiographs based on the given x-ray.
- The LLM (Vicuna) is fine-tuned on medical data (100k real conversations between patients and doctors) and ~30k radiology conversations to acquire domain specific and relevant features.
- We generate interactive and clean summaries (~217k) from free-text radiology reports of two datasets (MIMIC-CXR and OpenI). These summaries serve to enhance the performance of LLMs through fine-tuning the linear transformation layer on high-quality data. For more details regarding our high-quality summaries, please check Dataset Creation.
- We align frozen medical visual encoder (MedClip) with a fune-tuned LLM (Vicuna), using simple linear transformation.
1. Prepare the code and the environment
Clone the repository and create a anaconda environment
git clone https://github.com/mbzuai-oryx/XrayGPT.git
cd XrayGPT
conda env create -f env.yml
conda activate xraygptOR
git clone https://github.com/mbzuai-oryx/XrayGPT.git
cd XrayGPT
conda create -n xraygpt python=3.9
conda activate xraygpt
pip install -r xraygpt_requirements.txt1. Prepare the Datasets for training
Refer the dataset_creation for more details.
Download the preprocessed annoatations mimic & openi. Respective image folders contains the images from the dataset.
Following will be the final dataset folder structure:
dataset
├── mimic
| ├── image
| | ├──abea5eb9-b7c32823-3a14c5ca-77868030-69c83139.jpg
| | ├──427446c1-881f5cce-85191ce1-91a58ba9-0a57d3f5.jpg
| | .....
| ├──filter_cap.json
├── openi
| ├── image
| | ├──1.jpg
| | ├──2.jpg
| | .....
| ├──filter_cap.json
...
3. Prepare the pretrained Vicuna weights
We built XrayGPT on the v1 versoin of Vicuna-7B. We finetuned Vicuna using curated radiology report samples. Download the Vicuna weights from vicuna_weights The final weights would be in a single folder in a structure similar to the following:
vicuna_weights
├── config.json
├── generation_config.json
├── pytorch_model.bin.index.json
├── pytorch_model-00001-of-00003.bin
...
Then, set the path to the vicuna weight in the model config file "xraygpt/configs/models/xraygpt.yaml" at Line 16.
To finetune Vicuna on radiology samples please download our curated radiology and medical_healthcare conversational samples and refer the original Vicuna repo for finetune.Vicuna_Finetune
4. Download the pretrained Minigpt-4 checkpoint
Download the pretrained minigpt-4 checkpoints. ckpt
A. First mimic pretraining stage
In the first pretrained stage, the model is trained using image-text pairs from preprocessed mimic dataset.
To launch the first stage training, run the following command. In our experiments, we use 4 AMD MI250X GPUs.
torchrun --nproc-per-node NUM_GPU train.py --cfg-path train_configs/xraygpt_mimic_pretrain.yaml2. Second openi finetuning stage
In the second stage, we use a small high quality image-text pair openi dataset preprocessed by us.
Run the following command. In our experiments, we use AMD MI250X GPU.
torchrun --nproc-per-node NUM_GPU train.py --cfg-path train_configs/xraygpt_openi_finetune.yamlDownload the pretrained xraygpt checkpoints. link
Add this ckpt in "eval_configs/xraygpt_eval.yaml".
Try gradio demo.py on your local machine with following
python demo.py --cfg-path eval_configs/xraygpt_eval.yaml --gpu-id 0
 |
 |
 |
 |
- MiniGPT-4 Enhancing Vision-language Understanding with Advanced Large Language Models. We built our model on top of MiniGPT-4.
- MedCLIP Contrastive Learning from Unpaired Medical Images and Texts. We used medical aware image encoder from MedCLIP.
- BLIP2 The model architecture of XrayGPT follows BLIP-2.
- Lavis This repository is built upon Lavis!
- Vicuna The fantastic language ability of Vicuna is just amazing. And it is open-source!
If you're using XrayGPT in your research or applications, please cite using this BibTeX:
@article{Omkar2023XrayGPT,
title={XrayGPT: Chest Radiographs Summarization using Large Medical Vision-Language Models},
author={Omkar Thawkar, Abdelrahman Shaker, Sahal Shaji Mullappilly, Hisham Cholakkal, Rao Muhammad Anwer, Salman Khan, Jorma Laaksonen and Fahad Shahbaz Khan},
journal={arXiv: 2306.07971},
year={2023}
}This repository is licensed under CC BY-NC-SA. Please refer to the license terms here.


