Ryan Eckert -- ryan.j.eckert@gmail.com
This repository contains scripts and data associated with the publication:
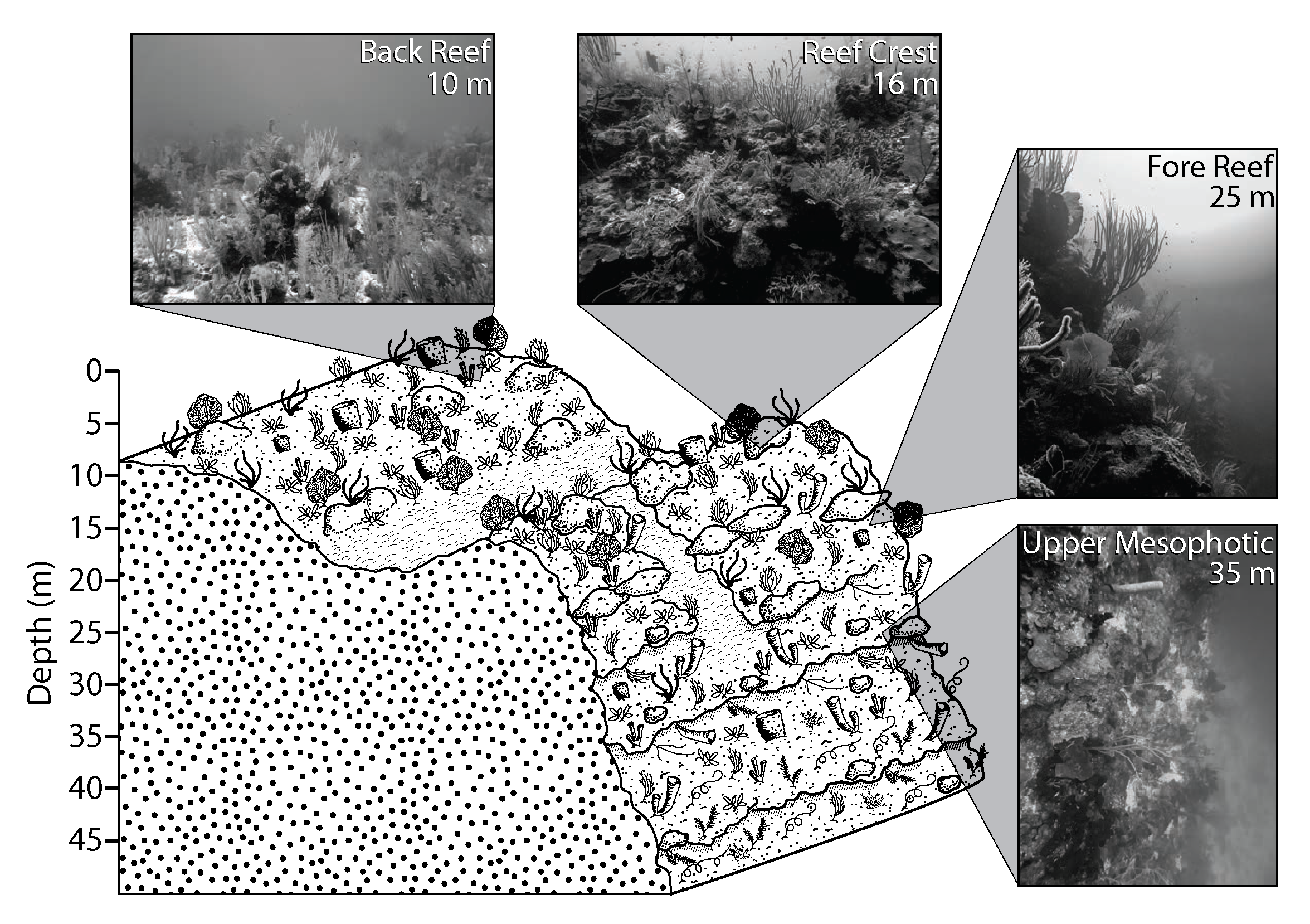
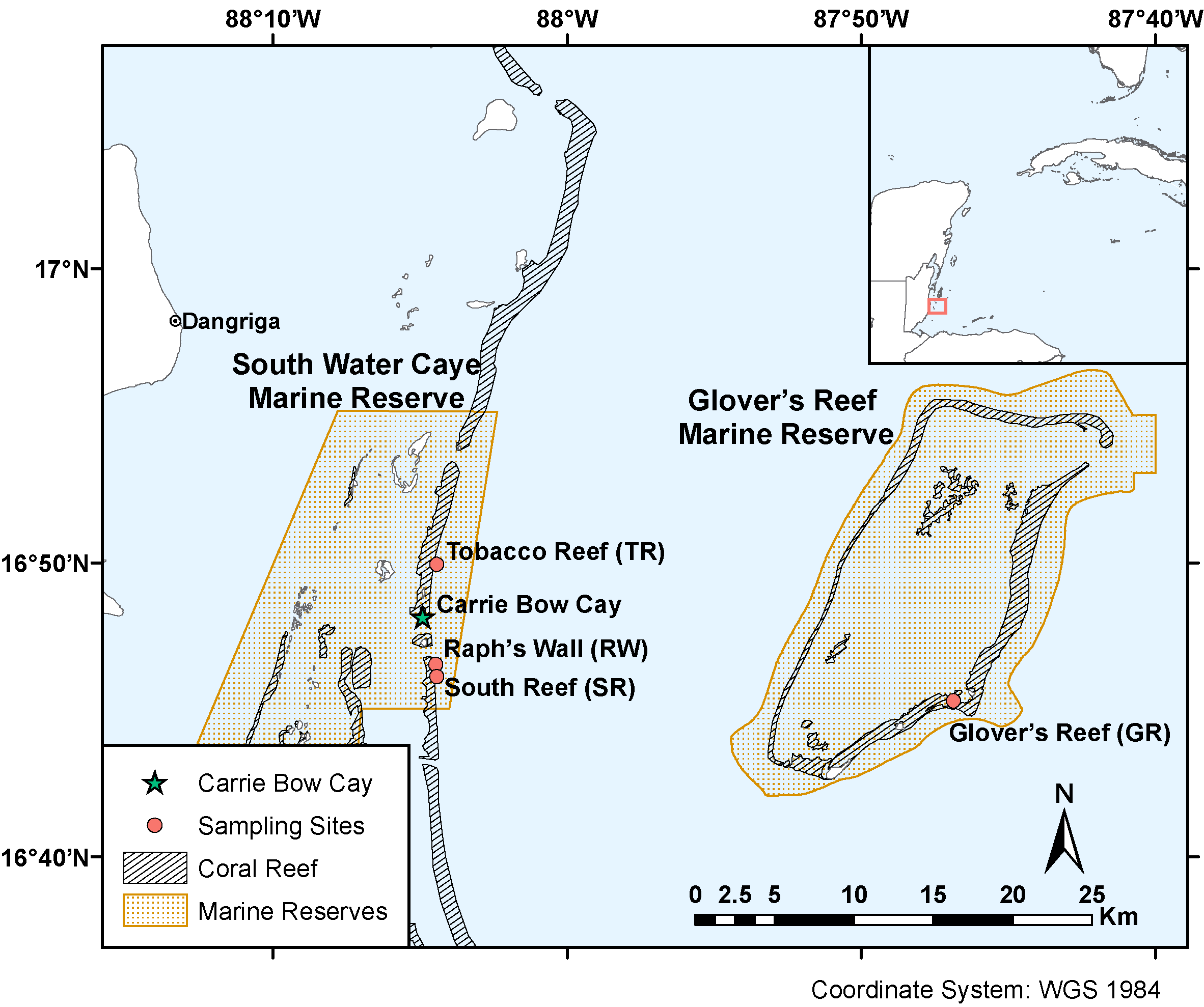
In Belize, shallow populations (10 and 16 m) of the coral species Montastraea cavernosa from the back reef and reef crest are genetically differentiated from deeper populations on the fore reef and reef wall (25 and 35 m). Like many species of scleractinian corals, M. cavernosa has an obligate symbiosis with dinoflagellate microalgae from the family Symbiodiniaceae. Here, we describe the Symbiodiniaceae taxa found within previously-sampled and genotyped M. cavernosa populations along a depth gradient on the Belize Barrier Reef by implementing high-throughput sequencing of the ITS2 region of Symbiodiniaceae ribosomal DNA and the SymPortal analysis framework. While Symbiodiniaceae ITS2 type profiles across all sampling depths were almost entirely (99.99%) from the genus Cladocopium (formerly Symbiodinium Clade C), shallow (10 and 16 m) populations had a greater diversity of ITS2 type profiles in comparison to deeper (25 and 35 m) populations. Permutational multivariate analysis of variance (PERMANOVA) confirmed significant differences in ITS2 type profiles between shallow and deep sample populations. Overall Symbiodiniaceae communities changed significantly with depth, following patterns similar to the coral host’s population genetic structure. Though physiological differences among species in the cosmopolitan genus Cladocopium are not well-described, our results suggest that although some members of Cladocopium are depth-generalists, shallow M. cavernosa populations in Belize may harbor shallow-specialized Symbiodiniaceae not found in deeper populations.
Lab protocols adapted from Meiog et al. 2009; Klepac et al. 2013.
-
figures/
- Fig1.eps -- Diagramatic representation of sampling depths (.eps)
- Fig1.png -- Diagramatic representation of sampling depths (.png)
- Fig2.eps -- Map of sampling sites (.eps)
- Fig2.png -- Map of sampling sites (.png)
- Fig3.eps -- Barplot of ITS2 sequences
- Fig4.eps -- Barplot of ITS2 type profiles
- Fig5.eps -- nMDS plots of ITS2 sequences and ITS2 type profiles
-
lab_protocol/
- barcodeMM.csv -- Barcoding PCR mastermix recipe
- bcPCR.csv -- Barcoding PCR profile
- bcPrimers.csv -- Barcoding PCR primer sequence example
- ctab.csv -- CTAB extraction buffer recipe
- etbrGel.csv -- Ethidium bromide gel recipe
- index.html -- Symbiodiniaceae lab prorocol webpage
- its2MM.csv -- Symbiodiniaceae ITS2 mastermix recipe
- its2PCR.csv -- Symbiodiniaceae ITS2 PCR profile
- its2Primers.csv -- Symbiodiniaceae specific ITS2 primer sequences
- reagents.csv -- Reagent stock recipes
- supplies.csv -- Supplies for CTAB extraction
- sybrGel.csv -- SYBR gel recipe
-
scripts/
- sampleRename.py -- rename files based on a .csv table
-
stats/
- 62_20190310_DBV_2019-03-11_01-11-25.167036.profiles.absolute.clean.txt -- SymPortal ITS2 type profile absolute abundance output file cleaned for importing into R
- 62_20190310_DBV_2019-03-11_01-11-25.167036.profiles.absolute.txt -- SymPortal ITS2 type profile absolute abundance output file
- 62_20190310_DBV_2019-03-11_01-11-25.167036.profiles.relative.txt -- SymPortal ITS2 type profile relative abundance output file
- 62_20190310_DBV_2019-03-11_01-11-25.167036.seqs.absolute.clean.txt -- SymPortal ITS2 sequence absolute abundance output file cleaned for importing into R
- 62_20190310_DBV_2019-03-11_01-11-25.167036.seqs.absolute.txt -- SymPortal ITS2 sequence absolute abundance output file
- 62_20190310_DBV_2019-03-11_01-11-25.167036.seqs.relative.txt -- SymPortal ITS2 sequence relative abundance output file
- CBC_MCAV_sampling_metadata.txt -- M. cavernosa sample metadata
- Symbiodiniaceae_ITS2_statistical_analyses.Rmd -- Symbiodiniaceae statistical analysis Rmarkdown document
- index.html -- Symbiodiniaceae statistical analysis webpage
- its2Primer.pwk -- PRIMER7 workbook
- stats.Rproj -- R project file
-
.gitignore -- .gitignore file
-
CBC_MCAV_Symbiodiniaceae_ITS2_metadata.xlsx -- M. cavernosa sample and library prep metadata
-
README.md -- Repository readme document
-
Symbiodiniaceae_ITS2_primers_PCR.xlsx -- Symbiodiniaceae ITS2 and barcoding primer sequences and PCR profiles