DeepSEA is a model to predicting effects of noncoding variants. This is implemented by tensorflow-2.0 again.
CNN + Pool + CNN + Pool + CNN + Dense + Dense
Binary Cross Entropy
SGD with Momentum (momentum = 0.9)
We run the code on Ubuntu 18.04 LTS with a GTX 1080ti GPU.
Python (3.7.3) | Tensorflow (2.0.0) | CUDA (10.0) | cuDNN (7.6.0)
You need to first download the training, validation, and testing sets from DeepSEA. You can download the datasets from
here. After you have extracted the
contents of the tar.gz file, move the 3 .mat files into the ./data/ folder.
The model that trained by myself is available in BAIDU Net Disk here
Because of my RAM limited, I firstly transform the train.mat file to .tfrecord files.
python preprocess.py
Then you can train the model initially.
CUDA_VISIBLE_DEVICES=0 python main.py -e train
When you have trained successfully, you can evaluate the model.
CUDA_VISIBLE_DEVICES=0 python main.py -e test
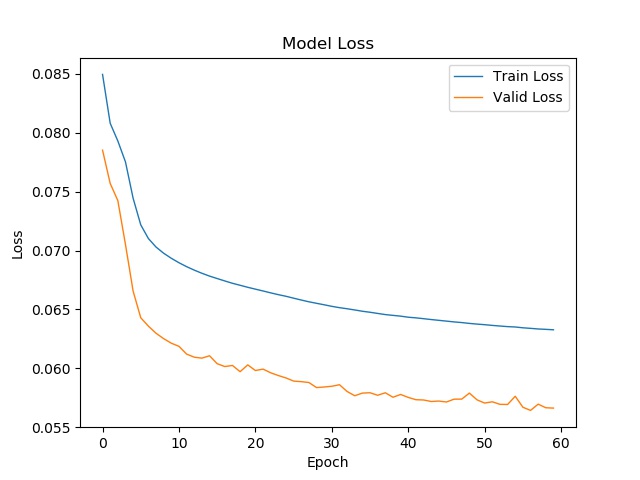
You can get my result in the ./result/ directory.
We use two metric to evaluate the model. (AUROC, AUPR)
| - | DNase | TFBinding | HistoneMark | All |
|---|---|---|---|---|
| AUROC | 0.8925 | 0.9071 | 0.8270 | 0.8960 |
| AUPR | 0.3809 | 0.2400 | 0.3277 | 0.2691 |
Predicting effects of noncoding variants with deep learning-based sequence model | Github