We provide a docker setup with all dependencies and runtime configuration. To start the container follow:
- Have installed:
- docker (>=20.0.0)
- docker compose (>=2.0.0!, notice, that the page suggests installing older version)
- additionally, to run using GPU:
- cuda (>=11.3) , nvcc and cudnn (>= 8.0)
- nvidia docker runtime
- Clone this repository.
sudo chmod 744 ./start.sh ./stop.sh- Set the environment configuration in the
docker/.env:PYTORCH,CUDA,CUDNN- only for GPU, modify only if needed. Before changing verify against Minkowski Engine requirementsDATA_PATH- if your name is Adaś and you are computer you should leave it as it is. If your name is Ania, Witek or Konrad, RNG guesses it is../../data/. In other cases set the path to the folder containing data
- Start container using
./start.sh. If you want to run container on cpu use./start.sh cpu. - To stop use
./stop.shor./stop.sh cpufor cpu respectively.
- Mark
srcdirectory as the source root:- right click on
srcdirectory -> Mark Directory As -> Source root - with this step done, PyCharm will consider
srcas source directory, therefore to reachligands-classificationdirectory remember to use../in your code.
- right click on
- Exclude unnecessary directories from indexing:
- PyCharm automatically indexes files in the project, so that it can propose i.e. path completion, yet some directories should not be indexed:
- similarly to the first step: right click on directory -> Mark Directory As -> Excluded
- directories to exclude:
.neptune,dataandvenvif present
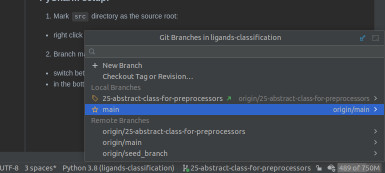
- Branch management:
- switch between the branches using PyCharm GUI, not with terminal tools
- in the bottom-right corner, there is a special menu:
- select the desired branch and choose
Checkoutin order to switch branches, - if you can't see a branch, make sure to run
git fetchfrom the Git toolbar menu.
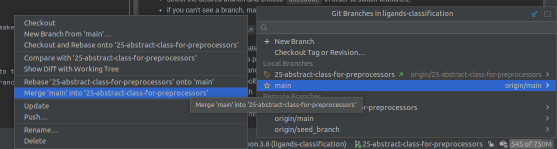
- Pulling changes from
mainbranch into a task branch:- should there be some update on the
mainbranch, please make sure to include them and resolve potential merge conflicts before creating a pull request from your task branch - checkout to the
mainbranch - update project using Git toolbar menu
- checkout to the task branch
- open branch menu -> choose main branch -> Merge 'main' into task branch
- thanks to the above, all commits merged into the
mainbranch will be applied to the specific branch
- should there be some update on the