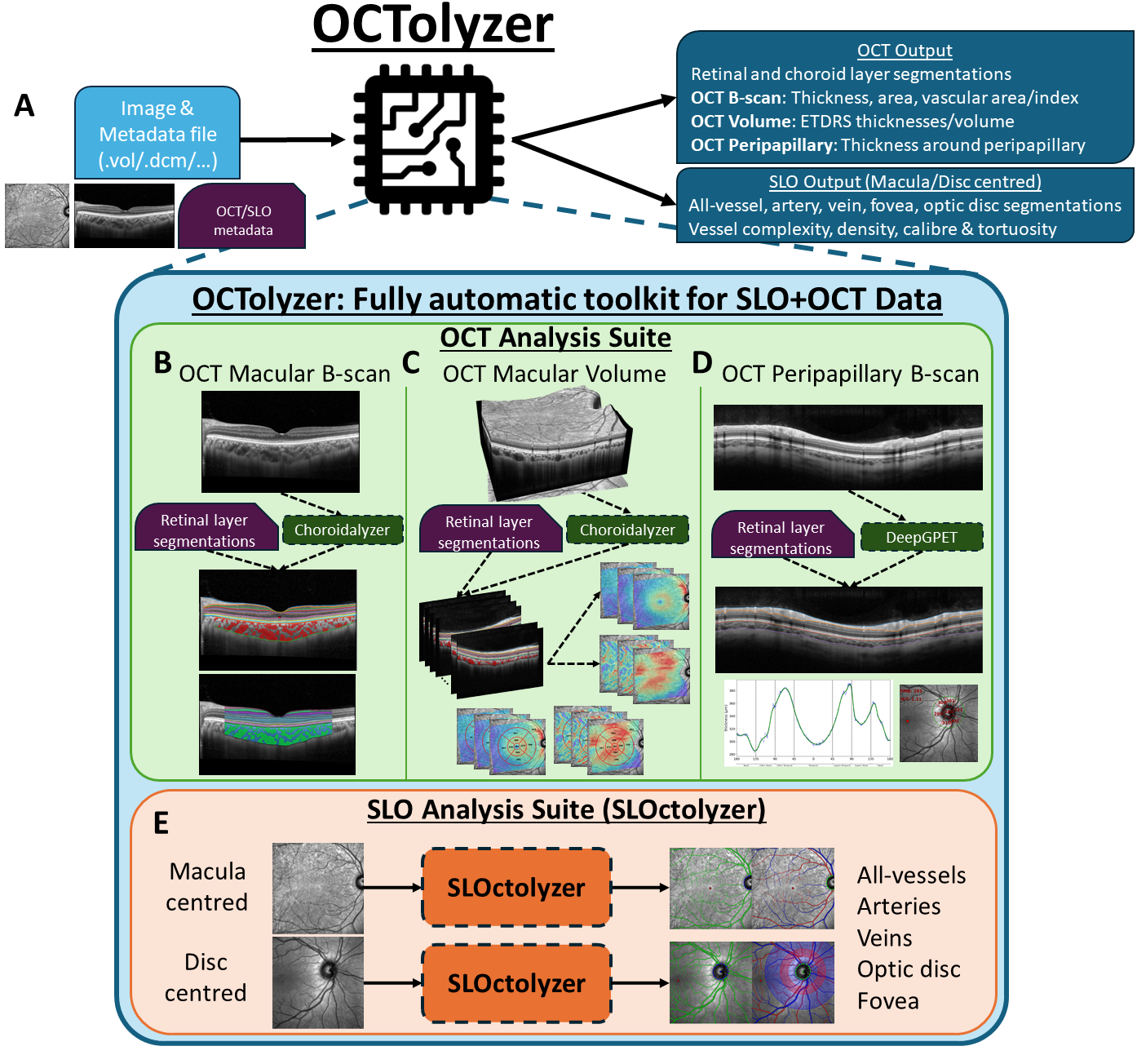
OCTolyzer: A fully automatic toolkit for segmentation and feature extracting in optical coherence tomography (OCT) and scanning laser ophthalmoscopy (SLO) data
OCTolyzer is a fully automatic pipeline capable of fully characterising the retinal and choroidal layers as seen on cross-sectional OCT images. It also supports segmentation and analysis of the en face retinal vessels on the corresponding confocal, infra-red scanning laser ophthalmoscopy (SLO) image, with additional detection of the fovea and optic disc. The pipeline utilises several fully automatic deep learning methods for segmentation of the OCT choroid and en face retinal vessels, and utilises the OCT retinal layer segmentations from the imaging device's built-in software, acessible from from file metadata, for generating retinochoroidal OCT measurements and en face retinal vessel measurements.
Please find the pre-print describing OCTolyzer's pipeline here, which is currently under review at "New Frontiers in Optical Coherence Tomography" special issue of ARVO's Translational Vision Science and Technology.
OCTolyzer is also capable of extracting clinically-relevant features of interest of the retina and choroid. The code used to measure regional and spatial measurements from OCT images was developed in-house, while the code used to measure features of en face retinal vessels were based on the code produced by Automorph, whose codebase can be found here.
See below for a visual description of OCTolyzer's analysis pipeline.
.
├── figures/ # Figures for README
├── instructions/ # Instructions, installation, manual annotation help
├── octolyzer/ # core module for carrying out segmentation and feature measurement
├── config.txt # Text file to specify options, such as the analysis directory path.
├── README.md # This file
└── usage.ipynb # Demonstrative Jupyter notebook to see usage.
- The code found in
octolyzer
.
├── octolyzer/
├───── measure/ # Feature extraction
├───── segment/ # Segmentation inference modules.
├───── __init__.py
├───── analyse.py # Script to segment and measure a single OCT+SLO .vol file and save out results. Can be used interactively via VSCode or Notebooks.
├───── analyse_slo.py # Script for SLO image analysis, adapted from SLOctolyzer.
├───── collate_data.py # Combine individual file results for collating into single, summary file for batch processing.
├───── main.py # Wrapper script to run OCTolyzer from the terminal for batch processing.
└───── utils.py # Utility functions for plotting and processing segmentations.
To get a local copy up follow the steps in instructions/quick_start.txt, or follow the instructions below.
-
Clone the OCTolyzer repository via
git clone https://github.com/jaburke166/OCTolyzer.git. -
You will need a local installation of python to run OCTolyzer. We recommend a lightweight package management system such as Miniconda. Follow the instructions here to download Miniconda for your desired operating system.
-
After downloading, navigate and open the Anaconda Prompt, and individually copy and run each line found in
install.txtto create your own environment in Miniconda and download necessary packages.- Note: if you have a GPU running locally to use OCTolyzer, line 3 in
instructions/install.txtshould bepip3 install torch torchvision --index-url https://download.pytorch.org/whl/cu121
- Note: if you have a GPU running locally to use OCTolyzer, line 3 in
Done! You have successfully set up the software to analyse SLO and OCT image data!
Now you can:
-
Launch notebooks using
jupyter notebookor jupyter labs usingjupyter laband see the minimal example below so that you can analyse your own SLO+OCT data. -
Alternatively, edit the
analysis_directoryandoutput_directoryoptions inconfig.txtand runpython path\to\OCTolyzer/octolyzer/main.pyto batch process your own OCT+SLO dataset.
If you have any problems using this toolkit, please do not hesitate to contact us - see the end of this README for contact details!
Please refer to usage.ipynb for an interactive demonstration of analysing SLO+OCT data.
# Load necessary modules
from octolyzer import utils, analyse
from pathlib import Path
# Detect images in analyze/demo
paths = sorted(Path("analyze/demo/").glob(f"*.vol"))
path = paths[0]
save_path = "analyze/output"
# Analyse SLO+OCT data - saved out in analyze/output/ into a folder whose name is the filename of the image
output = analyse.analyse(path, save_path)
# output is a tuple containing the results from the SLO analysis and OCT analysis separately
# Each tuple stores relevant (metadata, feature measurements, segmentations, logging)
At present, OCTolyzer only supports .vol files (native to Heidelberg Engineering), but there are plans in future to support more vendor neutral file formats such as .dcm.
We do not support .e2e files because current python-based file readers (EyePy, OCT-Converter) cannot locate the necessary pixel lengthscales to convert from pixel space to physical space. Moreover, we do not support regular image files because of the necessity to convert measurements from pixel space to physical space for standardised reporting of ocular measurements. Please do get in touch if you would like to help extend OCTolyzer on other file formats - see contact information at the end of this file!
See the documents in the instructions folder for details on installing and using OCTolyzer on your device.
Briefly, OCTolyzer can be run from the terminal using main.py for analysing batches of data, or using analyse.py individually per file using your favourite, interactive IDE such as VSCode or Jupyter Notebooks.
When using the terminal, you can specify certain input parameters using a configuration file, config.txt. Here, you can specify the analysis_directory and output_directory, i.e., where your data is stored and where you wish to save results to, respectively.
Currently, OCTolyzer supports fully automatic analysis of three .vol data types:
- Single, horizontal- or vertical-line macular OCT B-scans
- Posterior pole, macular OCT scans.
- Single, circular peripapillary OCT B-scans
These were the only OCT data types which the developers had access to, and thus could set up analysis streams for (after unpacking the files using EyePy). However, OCTolyzer is an evolving toolkit, so we expect other scan patterns to be supported if and when there is need for it. Check out octolyzer/utils.py for function load_volfile() to see how your scan pattern may be loaded in and supported and contact us!
At present, OCTolyzer is compatible with Windows and macOS operating systems. Given the compatibility with macOS, it's likely that Linux distributions will also work as expected, although has not been tested explicitly. The installation instructions are also the same across operating systems (once you have installed the relevant Miniconda Python distributions for your own operating system).
Once OCTolyzer is downloaded/cloned and the conda environment/python packages have been installed using the commands in instructions/install.txt, it is only the file path structures to be aware of when switching between OS, i.e. in the configuration file, config.txt, the analysis_directory and output_directory should be compatible with your OS.
OCTolyzer can run reasonably fast using a standard, GPU-less Windows laptop CPU. For only running the OCT analysis suite (i.e., analyse_slo set to 0).
- ~2 seconds for a single line OCT B-scan (pixel resolution
$496 \times 768)$ . - ~85 seconds for an OCT volume with thickness maps computed and measured for every retinochoroidal layer (pixel resolution
$61 \times 496 \times 768$ ). - ~3 seconds for an OCT peripapillary B-scan (pixel resolution
$768 \times 1536)$ . Note: To align the peripapillary grid, the localiser SLO image must be available, and the fovea and optic disc must be segmented, which increases execution time to around 30 seconds. If the SLO is unavailable, the alignment of the peripapillary grid is likely to off-centre from the fovea.
Note: Execution time increases when the SLO analysis suite is toggled on (analyse_slo set to 1), particularly for disc-centred SLO images (OCT peripapillary scans), because there are three regions of interest measured for each vessel type (all-vessel, artery, veins).
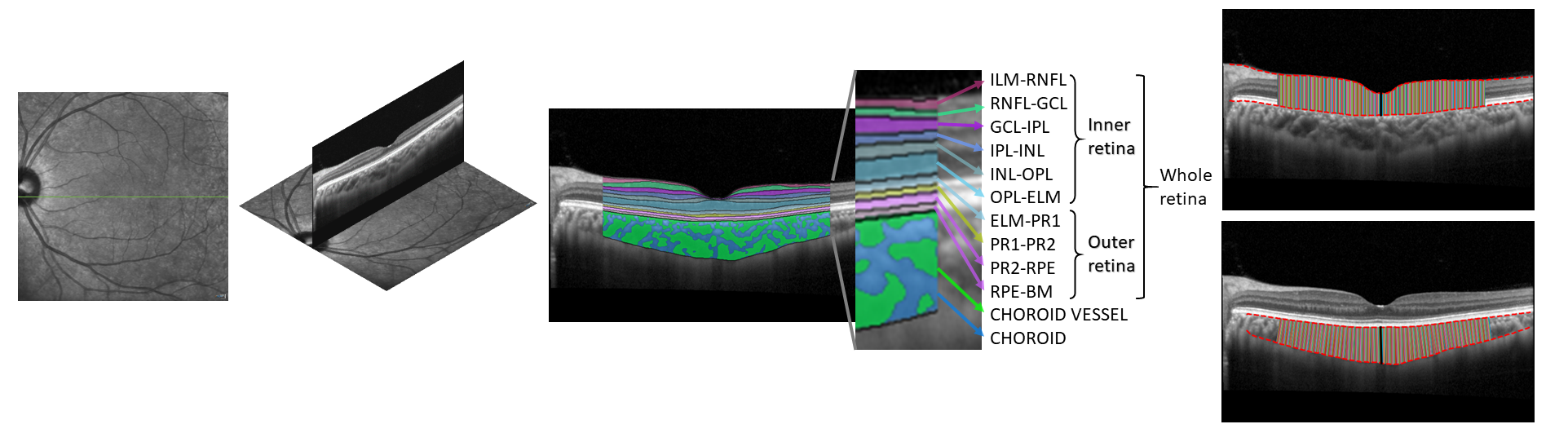
We do not provide functionality (yet) for automatic OCT retinal layer segmentation, but the automatic layer segmentations provided by Heidelberg's built-in software can be accessed directly from the .vol metadata for downstream measurements.
We use Choroidalyzer for OCT choroid segmentation of macular OCT scans (single-line B-scans and posterior pole scans) and DeepGPET for OCT choroid segmentation of peripapillary OCT B-scans. Choroidalyzer segments the choroidal space, vessels and detects the fovea on OCT B-scans, while DeepGPET only segments the choroidal space on the B-scan. Note: Choroidal vessels are not segmented on peripapillary B-scans currently. Please see the individual papers describing Choroidalyzer and DeepGPET's deep learning-based models here and here, respectively.
For SLO images, we have three separate deep learning-based models. One for binary vessel segmentation of the en face retinal vessels, and another for segmentation of the en face retinal vessels into arteries and veins, and simultaneous detection of the optic disc. A final, third model only detects on the fovea on the en face SLO image. Please see the separate pre-print for OCTolyzer's SLO analysis suite, SLOctolyzer. SLOctolzyer is also a stand-alone image analysis pipeline for SLO images (supporting both .vol and regular image files), and can be accessed and used freely here.
At present, OCTolyzer provides a suite of measurements for each of the three OCT data types which it supports:
Given some region of interest around the fovea (dictated by linescan_roi_distance in config.txt as a micron value around the fovea), area (in mm$^2$), average thickness and subfoveal thickness (in $\mu$m) will be measured for every available layer. Additionally for the choroid, vessel area and vascular index will also be measured and outputted. See below for a visualisation taken from the paper:
A note on choroid measurements: On occasion the choroid may appear skewed relative to the image axis, and this curvature can be accounted for by defining the the region of interest according to the choroid axis. By default, measurements of the choroid are made locally perpendicular to this axis with choroid_measure_type set to perpendicular. If this option is set to vertical, then the choroid is measured corresponding to the image axis, i.e. per A-scan (like the retina is, by default). In the above image, note the thickness measurements in the choroid are perpendicular to the upper boundary, but for the retina they are vertical. This option is only valid for macular OCT choroid image analysis, and not for peripapillary OCT choroid image analysis (as the local choroid curvature is due to the circular nature of the B-scan).
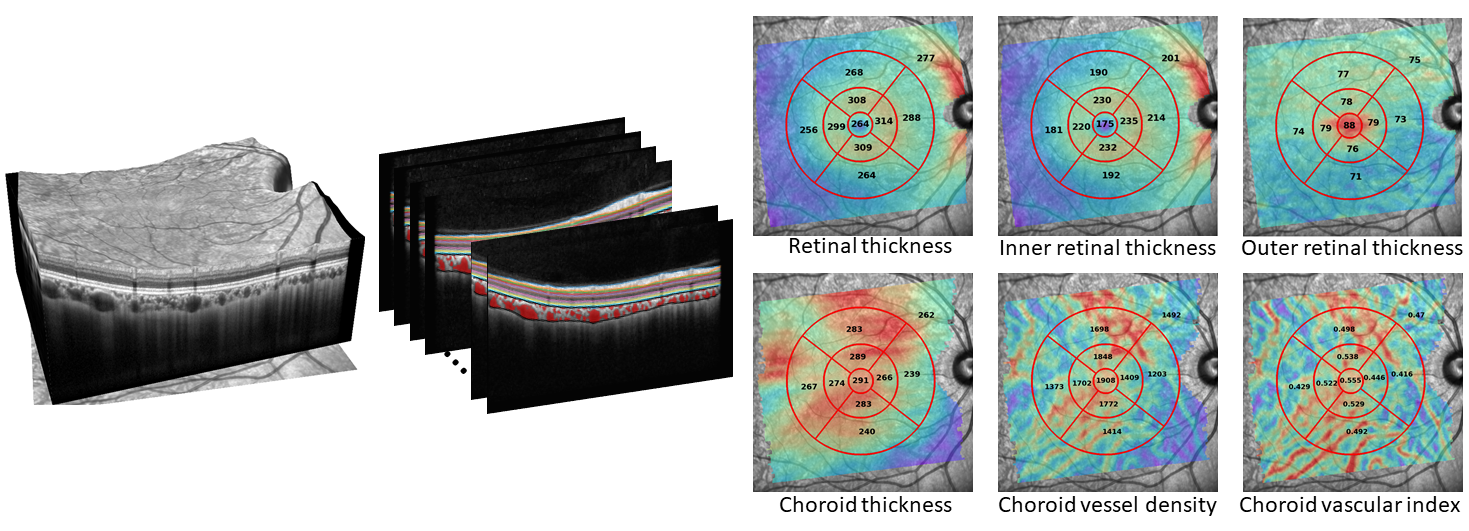
Thickness maps for all available retinal layers are generated, alongside choroid thickness, vessel density and vascular index maps. Volume maps are also generated for all metrics except for choroid vascular index.
Maps are then quantified by default using the ETDRS grid. Additional quantification of the map using an 8x8 posterior pole grid (7 mm square width) can also be done by setting analyse_square_grid to 1 in config.txt. If individual retinal layer segmentations are available, then you can specify analyse_all_maps to 1 to quantify all of these, or specify custom maps using the custom_maps option. See below for a visualisation taken from the paper:
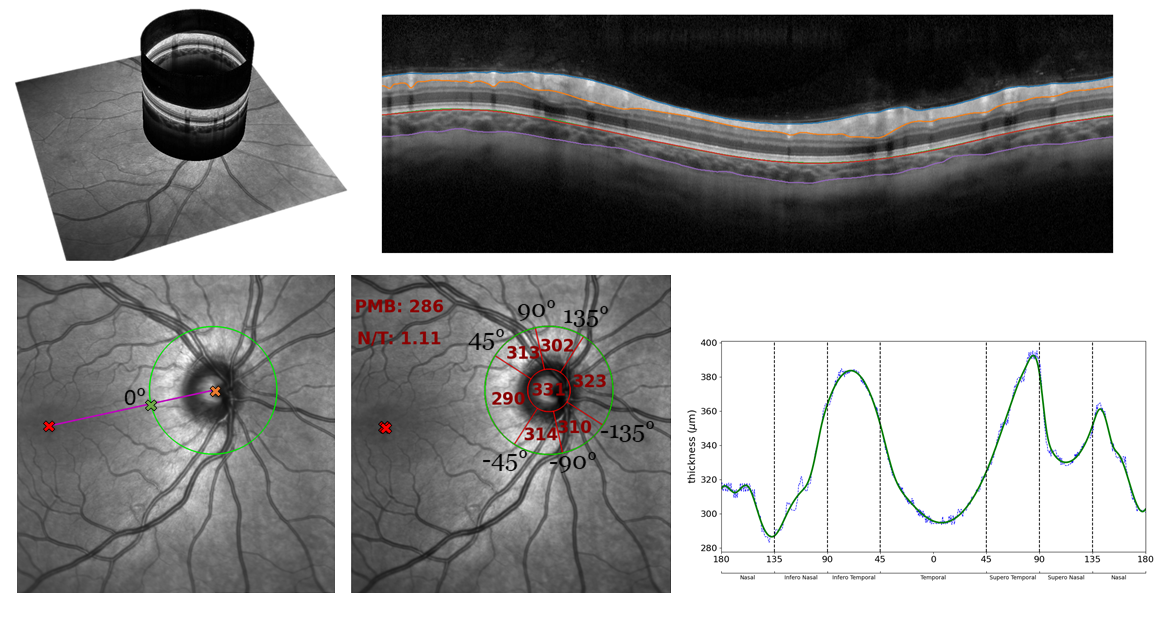
Average thickness in each of the six radial, peripapillary grids are measured, after aligning the centre of the temporal peripapillary grid with the detected fovea on the corresponding SLO image. To align the peripapillary grid properly, the localiser SLO image must be available. If the SLO is unavailable, the alignment of the peripapillary grid is likely to off-centre from the fovea. We also output the nasal-to-temporal ratio and average thickness in the papillomacular bundle. Thickness profiles around the disc are also quantified and saved out optionally. See below for a visualisation taken from the paper:
Note: Choroid analysis can be toggled off by setting analyse_choroid to 0 in config.txt.
At present, the features measured on the SLO image across the whole image are:
- Fractal dimension (dimensionless)
- Vessel perfusion density (dimensionless)
- Global vessel calibre (computed globally in microns or pixels, dependent on whether a conversion factor is specified, see below)
Additionally, for smaller regions of interest, zones B and C, and the whole image, the following measurements will be measured in pixels or microns, dependent on whether the conversion factor is specified (see below):
- Tortuosity density
- Local vessel calibre (similar to average vessel width, but computed and averaged across individual vessel segments)
- CRAE (for detected arteries only) using Knudston's formula
- CRVE (for detected veins only) using Knudston's formula
- arteriolar–venular ratio (AVR), which is CRAE divided by CRVE.
A note on tortuosity: A current limitation of the pipeline surrounds artery and vein map disconnectedness. Artery-vein crossings on the SLO are segmented such that only artery OR vein is classified, not both. This was an oversight at the point of ground truth annotation before model training. Thus, the individual artery/vein maps are disconnected. Unfortunately, this may have an impact on tortuosity density measurements.
Note: SLO analysis can be toggled off by setting analyse_slo to 0 in config.txt. This will reduce execution time.
We do not have any automatic functionality within OCTolyzer to correct any segmentation errors. Thus, we rely on the user to identify any visible problems with vessel classification.
However, we do provide functionality to manually correct en face retinal vessel segmentations on the SLO via ITK-Snap. There are instructions on using ITK-Snap for manual annotations in instructions/SLO_manual_annotations which describe how to setup ITK-Snap and use it to correct the binary vessel mask, and/or the artery-vein-optic disc segmentation masks.
Once the corrected segmentations are saved out as .nii.gz files in the same folder with the original .png segmentation mask(s), the pipeline can be run again and OCTolyzer should automatically identify these additional manual annotations and re-compute the features!
If you have any issues with running this toolkit on your own device, please contact us (see end of README for contact email).
This project and analysis toolkit is an evolving toolkit, so we are expecting unexpected errors to crop up on use-cases which the developers have not foreseen. We hope this pipeline will continue to be adapted to the needs of the end-user, so we welcome any and all feedback!
At the moment, setting robust_run to 1 will ensure that batch-processing does not fail if an unexpected error is caught. In fact, if an exception is found, details on the error in terms of it's type and full traceback are saved out in the process log (as well as printed out on the terminal/IDE) so the end-user may interrogate the codebase further and understand the source of the error.
If you are interested in only scanning laser ophthalmoscopy image analysis, check this repoistory out:
- SLOctolyzer: Analysis toolkit for automatic segmentation and measurement of retinal vessels on scanning laser ophthalmoscopy (SLO) images
If you are interested in automatic choroid analysis in OCT B-scans specifically, check these repositories out:
- Choroidalyzer: A fully automatic, deep learning-based tool for choroid region and vessel segmentation, and fovea detection in OCT B-scans.
- DeepGPET: fully automated choroid region segmentation in OCT B-scans.
- MMCQ: A semi-automatic algorithm for choroid vessel segmentation in OCT B-scans based on multi-scale quantisation, histogram equalisation and pixel clustering.
If you are interested in colour fundus photography (CFP) image analysis, check this repository out:
- Automorph: Automated retinal vascular morphology quantification via a deep learning pipeline.
The contributors to this method and codebase are:
- Jamie Burke (Jamie.Burke@ed.ac.uk)
If you wish to use this toolkit please consider citing our work using the following BibText
@article{burke2024octolyzer,
title={OCTolyzer: Fully automatic analysis toolkit for segmentation and feature extracting in optical coherence tomography (OCT) and scanning laser ophthalmoscopy (SLO) data},
author={Burke, Jamie and Engelmann, Justin and Gibbon, Samuel and Hamid, Charlene and Moukaddem, Diana and Pugh, Dan and Farrah, Tariq and Strang, Niall and Dhaun, Neeraj and MacGillivray, Tom and others},
journal={arXiv preprint arXiv:2407.14128},
year={2024}
}