The goal of circIMPACT is to detect the molecular pathways associated with the expression levels of certain circRNAs.
You can install the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("AFBuratin/circIMPACT")This is a basic example which shows you how to detect circRNA-impacted:
library(circIMPACT)
library(DESeq2)
library(data.table)
library(plyr)
library(Rtsne)
library(ggplot2)
library(ggrepel)
library(plotly)
library(ComplexHeatmap)
library(circlize)
library(viridis)
library(knitr)
library(kableExtra)
library(formattable)
library(htmltools)
library(sparkline)
library(tidyverse)
library(RColorBrewer)
library(purrr)
library(magrittr)
library(webshot)## load data for example
data("circularData")
data("meta")
data("coldata.df")
data("dds.circular")
## Filter out low circRNA
data.filt <- circularData[rowSums(circularData >= 5) >= 3,]
dds.filt.expr <- suppressMessages(DESeqDataSetFromMatrix(countData = ceiling(data.filt[,coldata.df$sample[order(coldata.df$condition)]]),
colData = coldata.df[order(coldata.df$condition),],
design = ~ condition))
dds.filt.expr <- suppressMessages(estimateSizeFactors(dds.filt.expr))
sf.filt <- sizeFactors(dds.filt.expr)
circNormDeseq <- counts(dds.filt.expr, normalized = T)Use marker.selection function to find out circRNA impacted specifing:
* adjusted p-value cutoff * log fold change cutoff * method for
calculate distance acrosso items * method for clustering
circIMPACT <- circIMPACT::marker.selection(dat = circNormDeseq,
dds = dds.filt.expr,
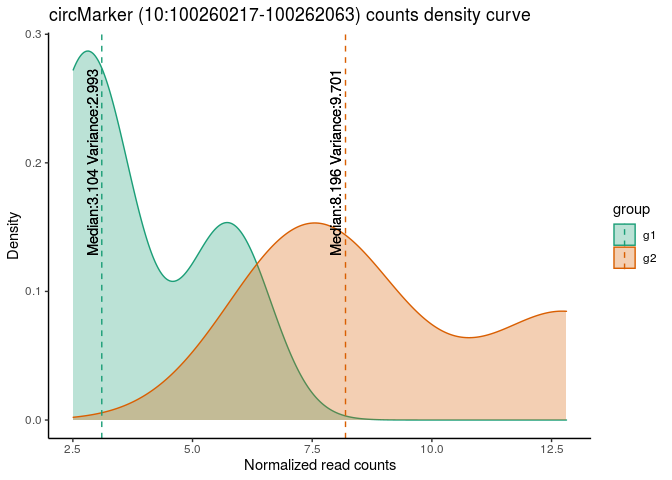
sf = sf.filt, p.cutoff = 0.05, lfc.cutoff = 1)For instance, you can see the distribution of circRNA-impact:
circMark <- circIMPACT$circ.targetIDS[1]
circMark_group.df <- circIMPACT$group.df[circIMPACT$group.df$circ_id==circMark,]
circMark_group.df$counts <- merge(circMark_group.df, reshape2::melt(circNormDeseq[circMark,]), by.x = "sample_id", by.y = "row.names")[,"value"]
mu <- ddply(circMark_group.df, "group", summarise, Mean=mean(counts), Median=median(counts), Variance=var(counts))
p <- ggplot(circMark_group.df, aes(x=counts, color=group, fill=group)) +
geom_density(alpha=0.3) +
geom_vline(data=mu, aes(xintercept=Median, color=group),
linetype="dashed") +
geom_text(data=mu, aes(x=Median[group=="g1"] - 0.2,
label=paste0("Median:", round(Median[group=="g1"], 3), " Variance:", round(Variance[group=="g1"], 3)), y=0.2),
colour="black", angle=90, text=element_text(size=9)) +
geom_text(data=mu, aes(x=Median[group=="g2"] - 0.2,
label=paste0("Median:", round(Median[group=="g2"], 3), " Variance:", round(Variance[group=="g2"], 3)), y=0.2),
colour="black", angle=90, text=element_text(size=11)) + scale_fill_brewer(palette="Dark2") +
scale_color_brewer(palette="Dark2") +
labs(title=paste0("circMarker (", circMark, ")", " counts density curve"), x = "Normalized read counts", y = "Density") +
theme_classic()
pYou’ll can use this list of circRNA to study a possible impact of their expression in genes deregulation.