brranching
Description
Brranching is an interface to many different sources of phylogenetic data (currently only from Phylomatic (http://phylodiversity.net/phylomatic/), but more sources to come) that allows users to query for phylogenetic data using taxonomic names.
Installation
Stable CRAN version
install.packages("brranching")Or dev version
install.packages("devtools")
devtools::install_github("ropensci/brranching")library("brranching")Phylomatic
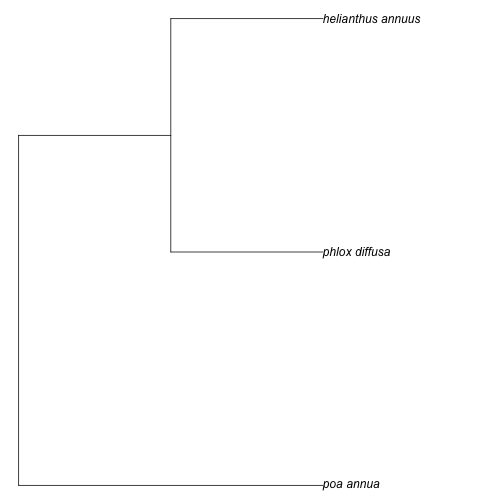
taxa <- c("Poa annua", "Phlox diffusa", "Helianthus annuus")
tree <- phylomatic(taxa=taxa, get = 'POST')
plot(tree, no.margin=TRUE)You can pass in up to about 5000 names. We can use taxize to get a random set of plant species names.
library("taxize")
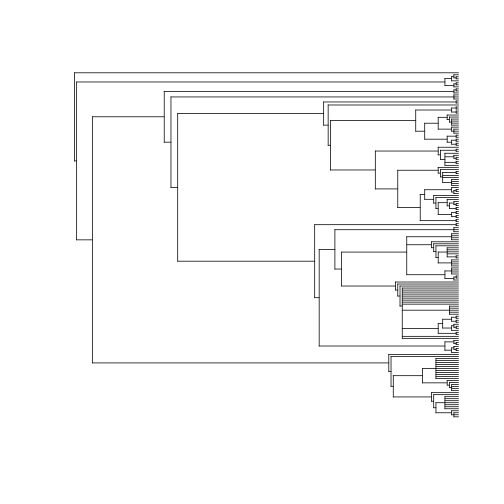
spp <- names_list("species", 200)
out <- phylomatic(taxa = spp, get = "POST")
plot(out, show.tip.label = FALSE)Meta
- Please report any issues or bugs.
- License: MIT
- Get citation information for
brranchingin R doingcitation(package = 'brranching') - Please note that this project is released with a Contributor Code of Conduct. By participating in this project you agree to abide by its terms.