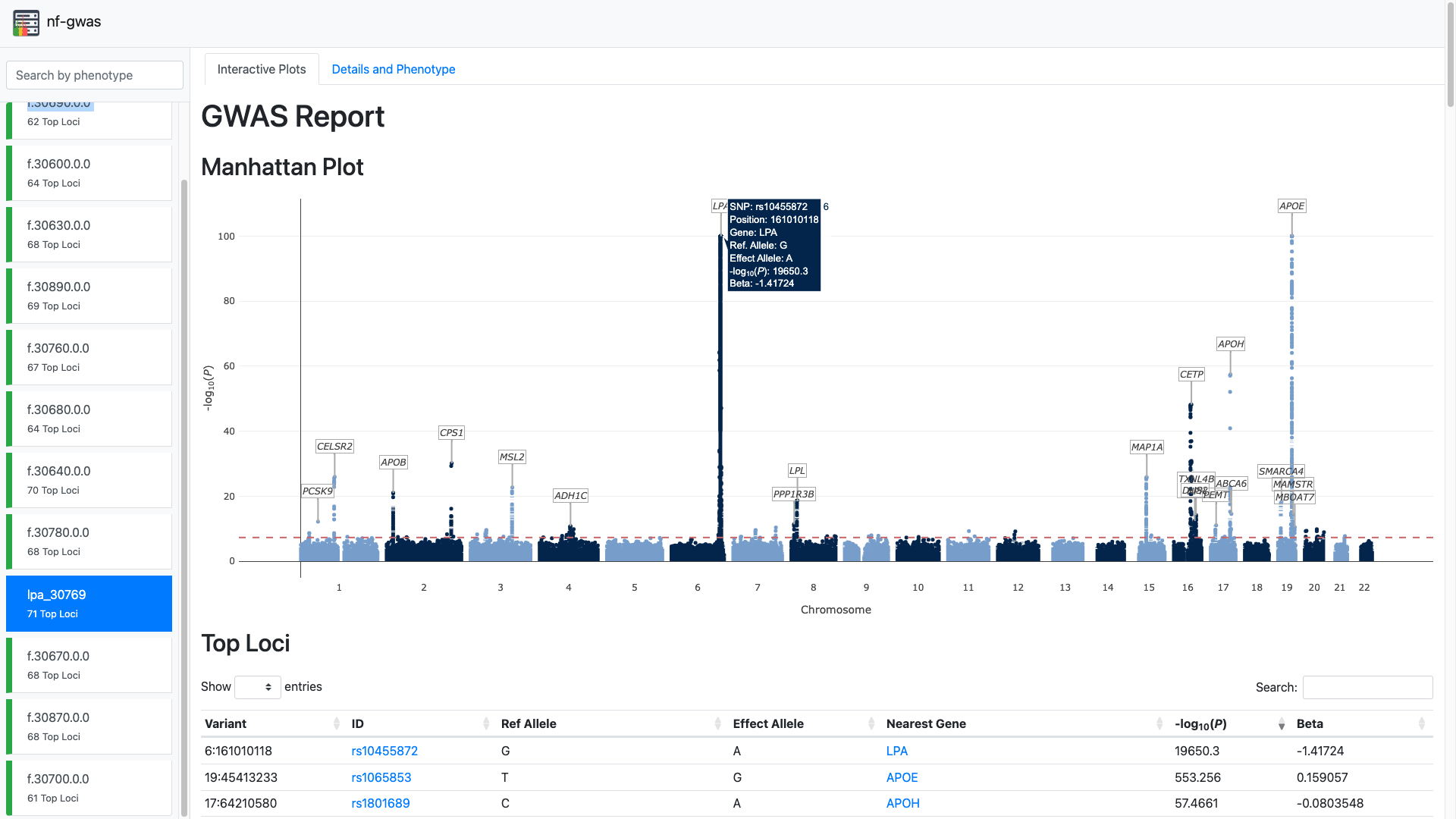
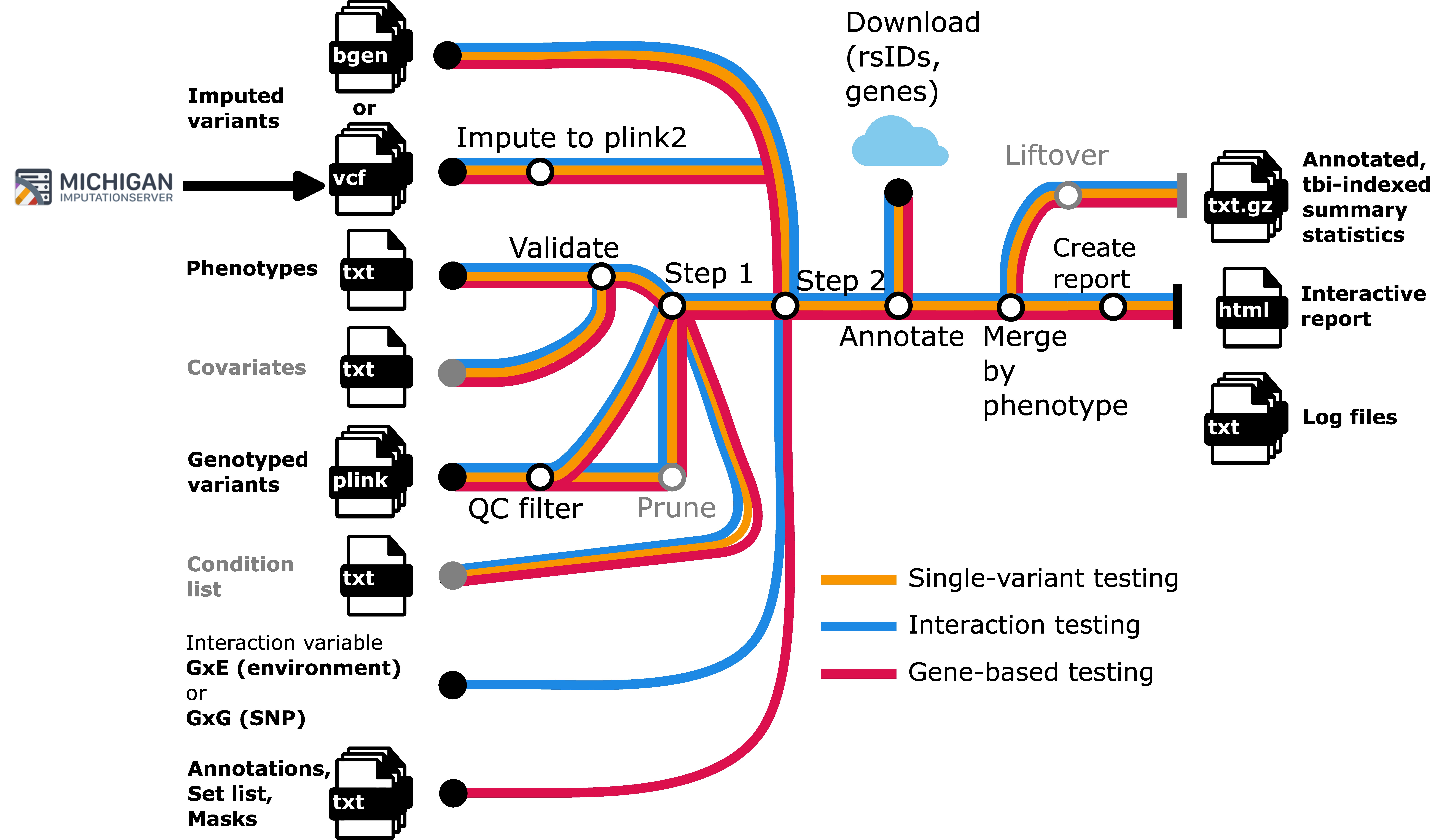
nf-gwas is a Nextflow pipeline to run biobank-scale genome-wide association studies (GWAS) analysis. The pipeline automatically performs numerous pre- and post-processing steps, integrates regression modeling from the REGENIE package and currently supports single-variant, gene-based and interaction testing. All modules are structured in sub-workflows which allows to extend the pipeline to other methods and tools in future. nf-gwas includes an extensive reporting functionality that allows to inspect thousands of phenotypes and navigate interactive Manhattan plots directly in the web browser.
The pipeline is tested using the unit-style testing framework nf-test and includes a schema definition to run with Nextflow Tower.
Documentation can be found here.
Please cite our paper if you use nf-gwas:
Schönherr S*, Schachtl-Riess JF*, Di Maio S*, Filosi M, Mark M, Lamina C, Fuchsberger C, Kronenberg F, Forer L. Performing highly parallelized and reproducible GWAS analysis on biobank-scale data. NAR Genom Bioinform. 2024 Feb 7;6(1):lqae015. doi: 10.1093/nargab/lqae015. PMID: 38327871; PMCID: PMC10849172.
-
Install Nextflow (>=22.10.4)
-
Run the pipeline on a test dataset
nextflow run genepi/nf-gwas -r v1.0.7 -profile test,<docker,singularity,slurm,slurm_with_scratch>
- Run the pipeline on your data
nextflow run genepi/nf-gwas -c <nextflow.config> -r v1.0.7 -profile <docker,singularity,slurm,slurm_with_scratch>
Please click here for available test config files.
git clone https://github.com/genepi/nf-gwas
cd nf-gwas
docker build -t genepi/nf-gwas . # don't ignore the dot
nextflow run main.nf -profile test,development
nf-gwas makes use of nf-test, a unit-style test framework for Nextflow.
cd nf-gwas
curl -fsSL https://code.askimed.com/install/nf-test | bash
./nf-test test
nf-gwas is MIT Licensed and was developed at the Institute of Genetic Epidemiology, Medical University of Innsbruck, Austria.