-
codon-msa: Contains scripts used in processing the reference data for codon aware multiple sequence alignment.
-
Viral_Sequence_Data: Contains inputs for the pipeline, such as the reference and genomes to be analyzed.
-
Intermediates: Contains intermediate files used in the pipeline.
-
MSA: Contains files used in aligning the genomes as well as data used when plotting final results.
-
tree: Contains the phylogenetic tree generated by FastTree.
-
hyphy_out: Contains final outputs for the pipeline such as a results summary and the final plot.
-
scripts: Contains the R script used to plot the data.
-
dag.svg: Representation of the overall workflow in SVG format. Viewable in web browser.
-
environment.yml: Contains dependencies used in this workflow. Used to build the conda environment.
-
Snakefile: Instructions for running the pipeline through snakemake. Can be configured to accept different inputs.
West Nile Virus is a mosquito-borne ssRNA (single stranded RNA) virus capable of causing encephalitis, meningitis, comas, and even death. Subsequent recovery from encephalitis caused by WNV is reportedly slow; less than 40% of patients report full recovery one year post-treatment [1]. Following the first reported case of WNV in the Western Hemisphere in 1999, the virus has spread throughout much of the United States and is a growing concern in Canada [2]. While current WNV incidence is relatively low, the amount of cases in steadily growing [3]. With increasing temperatures due to global climate change, specifically in Canada, the vector for this virus is spreading farther and farther north, increasing the risk of WNV spread and possible epidemic [4].
Positive selection analysis of viral genomes has been previously carried out on other viruses such as SARS-CoV-2 to identify viral adaptations and possible functional consequences of selected mutations [5]. Identification of the sites being selected for can help to characterize the changes the virus is undergoing as it further adapts to its new host: humans, and is a good starting point to identify mutations that may be involved in increased virality of this pathogen.
This pipeline will employ the FUBAR algorithm [6] to identify positively selected sites. This involves calculation and comparison of posterior probabilities of two rates: the rate of synonymous and non-synonymous mutations. Synonymous mutations comprise of mutations which do not affect the sequence of the coded protein, whereas non-synonymous mutations do. If the non-synonymous mutation rate (dN) is significantly greater than the synonymous mutation rate (dS), a site can be said to be experiencing positive (diversifying) selection.
This pipeline aims to identify what sites of West Nile Virus genomes are undergoing positive selection.
By identifying sites of positive selection, we may be able to identify factors related to the adaption of this virus. Positively-selected sites are candidates to analyze when looking for factors that may be related to virus survival and possibly, spread.
Installing this pipeline requires conda and git. Instructions for installing these
software can be found here and here respectively.
First, clone the repository by running the following command in a terminal:
git clone https://github.com/fieldima/biof501-pipeline.git
Next, build the conda environment by entering the following in the command line:
conda env create --file environment.yml
Now that it is installed, let's get to using it!
The pipeline is a relatively time-intensive process, depending on the amount of
sequences used, so I prefer to run it in a separate terminal window using tmux.
Before running the pipeline, always remember to activate the conda environment first
with the following command:
conda activate environment
The pipeline can then be run by typing in:
snakemake --cores 2
Notably, the number of cores can be changed by changing '2' to however many cores you wish to use.
A figure depicting the workflow is shown above. The workflow is broken up into 5 major steps.
-
Concatenation of Genomes with in-frame reference file. The input files of this pipeline include a Genomes file, containing the West Nile Virus genomes in a single file, and a reference file containing an in-frame coding sequence. This step combines the two files to prepare the data for selection analysis, which needs frame information.
-
Codon aware alignment. Preprocessing, msa, and postprocessing are all included in multiple sequence alignment of the genomes in a codon-preserving manner. Regular alignment procedures introduce frameshift artefacts into multiple sequence alignment data, which site-selection analysis cannot accept. This procedure converts genomes into amino acid format, forms a MSA of the genomes using
MAFFT, and finally converts the information back into DNA. -
Building the phylogenetic tree. A phylogenetic tree is inferred from MSA data using
FastTreeto determine evolutionary relationships between sampled West Nile Virus genomes. -
Positive selection analysis. The genomes are then analyzed using a FUBAR algorithm via
Hyphyto determine which sites are being selected for amongst all included genomes. -
Plotting analysis. Finally, the selection analysis is plotted via
Rusing thetidyverseandjsonlitepackages.
Two files are used as inputs; WNV_Genomes.fasta and Reference.fasta. These can be found in the Viral_Sequence_Data directory. WNV_Genomes.fasta includes all complete WNV genomes sampled in North America that are listed on NCBI Virus. Reference.fasta is an in-frame coding sequence for the genome.
The Genomes file can be altered to include or exclude different genomes to change the search space. For example, if I wanted to search for selected sites in European or Asian WNV variants, I would use a genomes file with only those sequences.
Changing the reference file can alter the scope of the analysis; e.g., if you wanted to determine if only a specific segment of the genome is under selection, like the envelope protein, you could change the reference file to only include that sequence.
Note: The reference file must be in frame for the pipeline to function correctly.
The four outputs of this pipeline are as follows:
-
A codon-aware alignment of the genomes. This file will be of type ".msa" and an example is provided in the MSA directory. This alignment file can be used in other analyses such as motif-identification or can be visualized itself using tools such as
IGV. Notably,Hyphyhas many other forms of analysis which make use of codon-aware alignments. -
A phylogenetic tree of the genomes. This will be of type "_tree" and an example is provided in the tree directory. This can be visualized by a plethora of different tools, such as the R packages
geiger,ggtree, andape. -
A summary of the selection analysis. This will be of type "_FUBAR" and an example is provided in the hyphy_out directory. This can be viewed with any text application, such as notepad in Windows or textedit in Mac. This will outline the analysis procedure as well as the most important results, i.e., the amount of sites with positive selection and the codon values.
-
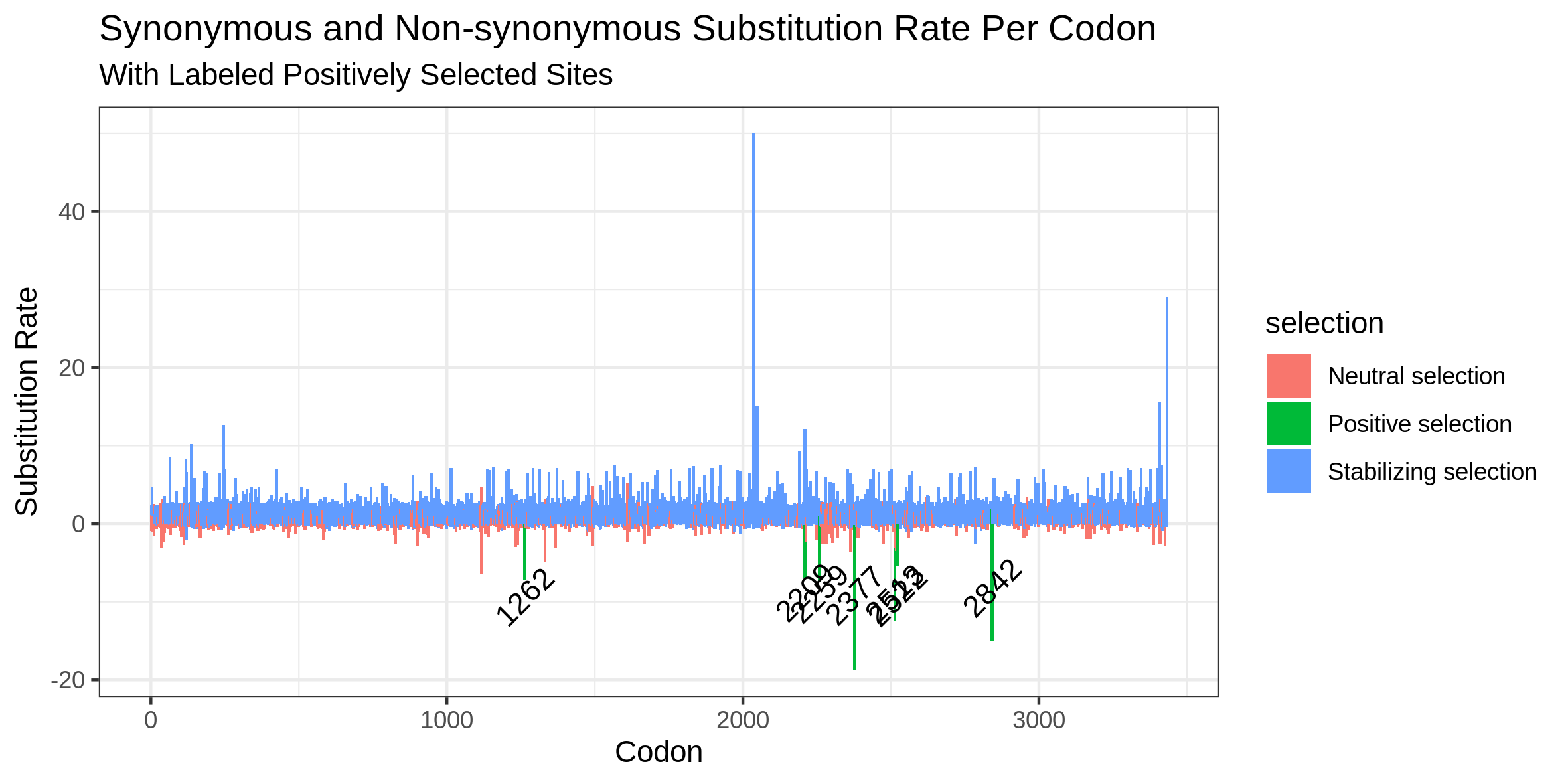
A plot depicting the analysis output. This will be of type ".png" and an example is provided in the hyphy_out directory. The x-axis represents the codon position, and the y-axis represents the substitution rate. Positive values represent the rate of synonymous substitution whereas negative values represent the non-synonymous rate. Sites of positive selection are labeled with their codon position for clarity.
Note: Example data are labeled as "Example" in their respective folders.
For West Nile Virus, 7 sites were found to be under positive selection. This is shown in the plot below.
The sites were mostly between codon 2200 and 2500, which corresponds to proteins NS4A and NS4B on the West Nile Virus genome.
Another notable site was codon 1262, which corresponds to NS2A.
As shown in the plot above, most sites were under purifying (negative), selection with some sites under neutral selection.
The finding that positive selection does occur on proteins NS4A and NS4B warrants a deeper analysis of the functional consequences of these mutations to identify possible reasons for selection. Some preliminary analysis could include GO term identification, functional domain annotation, etc. Another step of analysis would be to find which mutations are occurring on these proteins, to identify possible structural changes occurring in these proteins.
The pipeline itself could also be used to identify selection among different variants. It would be interesting to see the results of this pipeline if I used a different data set, like European or Asian WNV, to see if different areas are undergoing different selective pressures.
A modification of this pipeline could be to use a tool that contrasts different clades on this phylogeny, like MEME. That way, one could determine which sites are undergoing positive selection in one WNV variant over another. One such example could be contrasting selection between different host species of WNV. Do human carriers display different selection pressures than animal vectors?
Alternatively, this pipeline can be used to identify sites of positive selection in other viruses, such as COVID19, by changing the input files.
An improvement of this pipeline could be to automate the process of identifying which
proteins correspond to which codon positions in the Hyphy output. This would need to
incorporate gene reference data and coordinates.
[1] Campbell, G. L., Marfin, A. A., Lanciotti, R. S., & Gubler, D. J. (2002). West Nile virus. The Lancet Infectious Diseases, 2(9), 519-529. https://doi.org/10.1016/S1473-3099(02)00368-7
[2] Paz, S. (2015). Climate change impacts on West Nile virus transmission in a global context. Philosophical Transactions of the Royal Society B: Biological Sciences, 370(1665), 20130561. https://doi.org/10.1098/rstb.2013.0561
[3] Wijayasri, S., Nelder, M., Russell, C., Johnson, K., Johnson, S., Badiani, T., & Sider, D. (2019). West Nile virus illness in Ontario, Canada: 2017. Canada Communicable Disease Report, 44(1), 32-37. https://doi.org/10.14745/ccdr.v45i01a04
[4] Zheng, H., Drebot, M., & Coulthart, M. (2014). West Nile virus in Canada: ever-changing, but here to stay. Canada Communicable Disease Report, 40(10), 173-177. https://doi.org/10.14745/ccdr.v40i10a01
[5] Montoya, V., McLaughlin, A., Mordecai, G. J., Miller, R. L., & Joy, J. B. (2021). Variable routes to genomic and host adaptation among coronaviruses. Journal of Evolutionary Biology, 34(6), 924-936. https://doi.org/10.1111/jeb.13771
[6] Murrell, B., Moola, S., Mabona, A., Weighill, T., Sheward, D., Kosakovsky Pond, S. L., & Scheffler, K. (2013). FUBAR: A Fast, Unconstrained Bayesian AppRoximation for Inferring Selection. Molecular Biology and Evolution, 30(5), 1196-1205. https://doi.org/10.1093/molbev/mst030