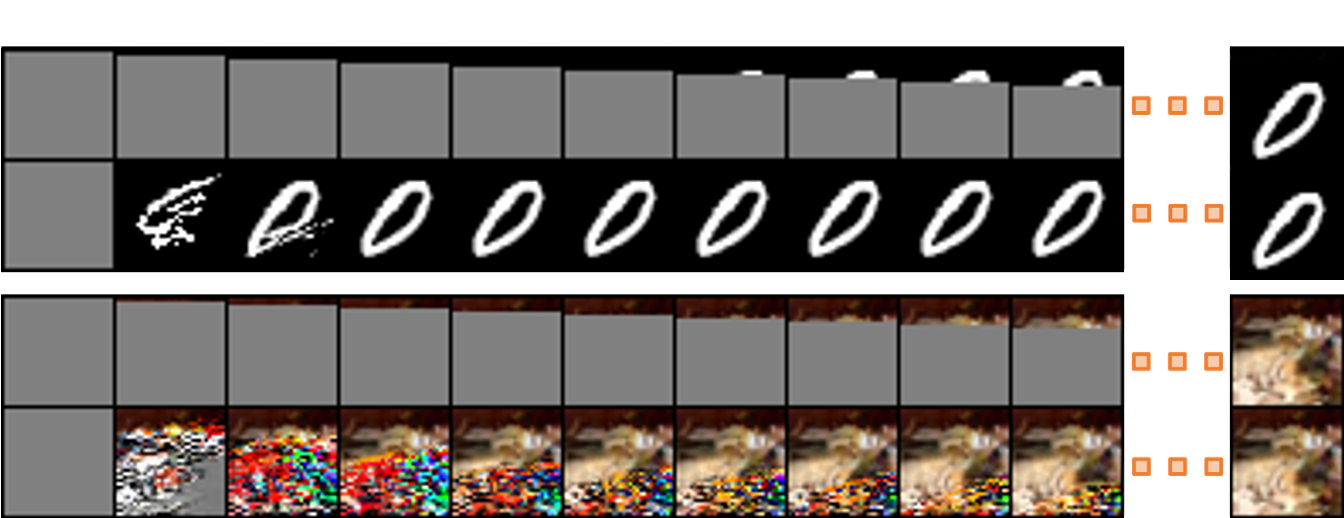This repo contains the official implementation for the ICML 2021 paper Accelerating Feedforward Computation via Parallel Nonlinear Equation Solving,
by Yang Song, Chenlin Meng, Renjie Liao, and Stefano Ermon.
We show that by viewing feedforward computation as a system of nonlinear equations, we can leverage parallel equation solvers to improve its speed. Our methods are particularly efficient when the computation graph contains many long skip connections, and can be used to accelerate the backpropagation of recurrent neural networks, inference of DenseNets, and autoregressive sampling from MADE and PixelCNN++.
jax==0.2.7
jaxlib==0.1.57+cuda101
flax==0.3.2
torch==1.7.1
torchvision==0.8.2
We run experiments by calling main.py with the following arguments.
usage: main.py [--runner RUNNER] [--config CONFIG]
arguments:
--runner RUNNER The runner to execute
--config CONFIG Path to the config filerunner and config can be specified below
| Experiment | --runner | --config |
|---|---|---|
| RNN backpropagation | BackpropRunner |
backprop.yml |
| DenseNet | DenseNetRunner |
densenet.yml |
| MADE sampling | MADESamplerRunner |
made_sampler.yml |
| PixelCNN++ sequential & Jacobi sampling | PixelCNNPPSamplerRunner |
pixelcnnpp_sampler.yml |
| PixelCNN++ sequential sampling w/ cache | CachedPixelCNNPPSamplerRunner |
cached_pixelcnnpp_sampler.yml |
| PixelCNN++ Jacobi-GS sampling | JacobiGSPixelCNNPPSamplerRunner |
jacobi_gs_pixelcnnpp_sampler.yml |
| PixelCNN++ GS-Jacobi sampling | GSJacobiPixelCNNPPSamplerRunner |
gs_jacobi_pixelcnnpp_sampler.yml |
The configurations of each experiment, such as dataset, block size, and algorithm, are provided in the corresponding config file, which should be straightforward to modify.
After running main.py, we can retrieve and plot experimental results by running *.ipynb files with Jupyter Notebook. We assume all experimental data are stored in folder plot_data/. Here is the correspondence between each experiment and the .ipynb file.
| Experiment | Jupyter Notebook file |
|---|---|
| RNN backpropagation | rnn_backprop.ipynb |
| DenseNet | densenet.ipynb |
| MADE sampling | made.ipynb |
| PixelCNN++ sampling | pixelcnnpp.ipynb |
We provide pretrained checkpoints (in subfolder runs/pretarined) and raw experimental data (in subfolder plot_data) via the following link:
https://drive.google.com/file/d/1cxV3pKUmLETt9veWRnEKt3SBCb-6sZDc/view?usp=sharing
If you find the idea or code useful for your research, please consider citing
@inproceedings{song2021accelerating,
title={Accelerating Feedforward Computation via Parallel Nonlinear Equation Solving},
author={Song, Yang and Meng, Chenlin and Liao, Renjie, and Ermon, Stefano},
booktitle = {International Conference on Machine Learning (ICML)},
year={2021},
}