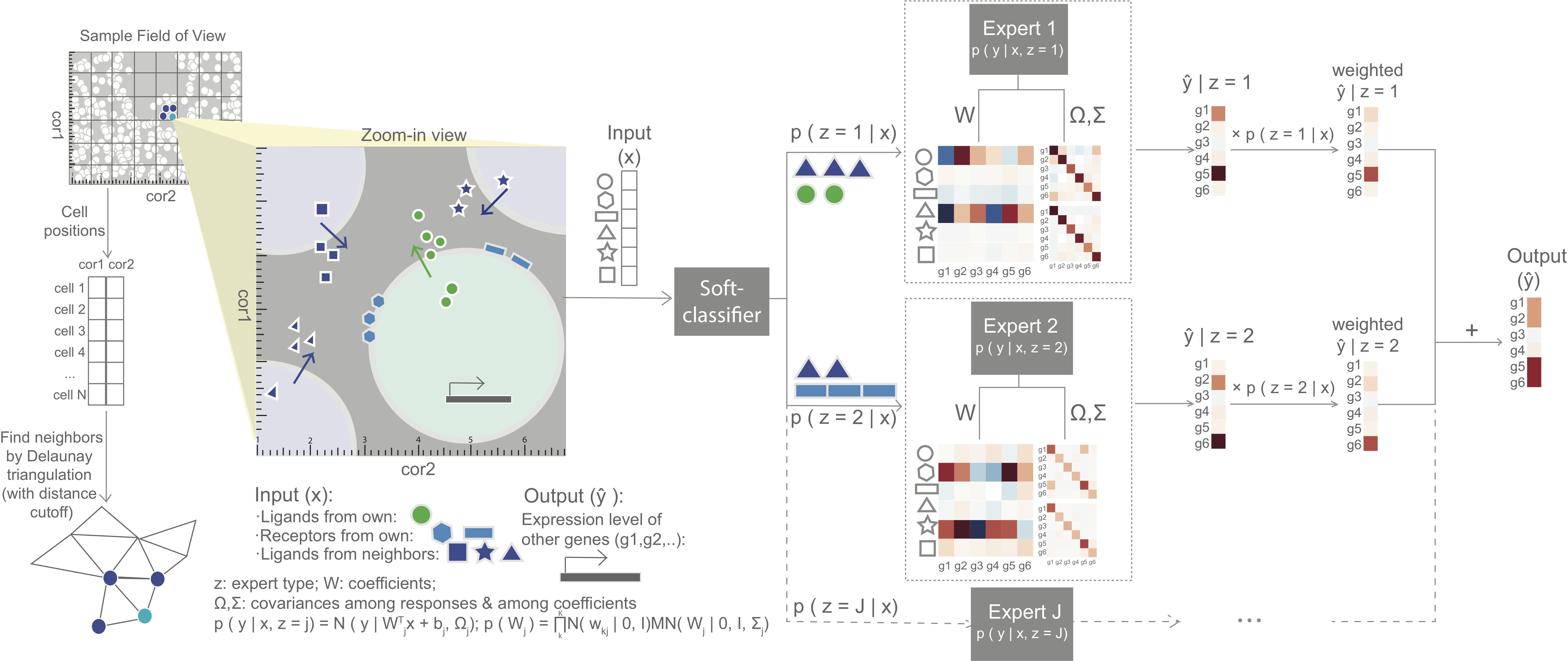
MESSI (Mixture of Experts for Spatial Signaling genes Identification) is a predictive framework to identify signaling genes active in cell-cell interaction. It jointly models gene interactions within and between cells, using the recently developed spatial single cell expression data. MESSI combines the ability to subdivide cell types with multi-task learning to accurately infer the expression of a set of response genes based on signaling genes and to provide useful biological insights about key signaling genes and cell subtypes.
- Python >= 3.6
- Python side-packages:
-- scikit-learn >= 0.22.1
-- scipy >= 1.3.0
-- numpy >= 1.16.3
-- pandas >= 0.25.3
It is recommended to use a virtural environment/pacakges manager such as Anaconda. After successfully installing Anaconda/Miniconda, create an environment by following:
conda create -n myenv python=3.6You can then install and run the package in the virtual environment. Activate the virtural environment by:
conda activate myenvMake sure you have pip installed in your environment. You may check by
conda listIf not installed, then:
conda install pipThen install MESSI, together with all its dependencies by:
pip install --upgrade https://github.com/doraadong/MESSI/tarball/masterIf you prefer not to use a virtual envrionment, then you may install MESSI and its dependencies by (may need to use sudo):
pip3 install --upgrade https://github.com/doraadong/MESSI/tarball/masterYou may find where the package is installed by:
pip show messiIn terminal, type (arguments are taken for example):
readyData.py -i ../input/ -d merfishThe usage of this script is listed as follows:
usage: readyData.py [-h] -i INPUT -d DATATYPE
optional arguments:
-h, --help show this help message and exit
-i INPUT, --input INPUT
string, path to the folder to save the expression
data, default 'input/'
-d DATATYPE, --dataType DATATYPE
string, type of expression data, default 'merfish'
Run MESSI by (arguments are taken for example):
messi -i ../input/ -o ../output/ -d merfish -g Female -b Parenting -c Excitatory -m train -c1 1 -c2 8 -e 5The usage of this file is listed as follows:
usage: messi [-h] -i INPUT [-ilr INPUT_LR] -o OUTPUT -d
{merfish,merfish_cell_line,starmap} -g GENDER -b BEHAVIOR -c
CELLTYPE -m MODE [-c1 NUMLEVEL1] [-c2 NUMLEVEL2] [-e EPOCHS]
[-gs GRID_SEARCH] [-ns N_SETS] [-r NUMREPLICATES] [-p PREPROCESS]
[-tr TOPKRESPONSES] [-ts TOPKSIGNALS]
optional arguments:
-h, --help show this help message and exit
-i INPUT, --input INPUT
string, path to the input folder with the expression
data, default 'input/'
-ilr INPUT_LR, --input_lr INPUT_LR
string, optional, path to the input folder with the
ligands and receptors list, default 'input/'
-o OUTPUT, --output OUTPUT
string, path to the output folder, default 'output/'
-d {merfish,merfish_cell_line,starmap}, --dataType {merfish,merfish_cell_line,starmap}
string, type of expression data, 'merfish' for MERFISH
hypothalamus data, 'merfish_cell_line' for MERFISH U-2
OS cells, 'starmap' for 'STARmap mPFC cells';default
'merfish'
-g GENDER, --gender GENDER
string, gender of input animal sample, default
'Female', put 'na' if not available
-b BEHAVIOR, --behavior BEHAVIOR
string, behavior of input animal sample, default
'Naive', put 'na' if not available
-c CELLTYPE, --cellType CELLTYPE
string, cell type that will be built a model for, use
\ for white-space, e.g. 'OD\ Mature\ 2', default
'Excitatory'
-m MODE, --mode MODE string, any of 'train', 'CV'; if 'train', then all
data will be used for training and output a pickle
file for learned parameters; if 'CV', then cross-
validation will be conducted each time with an
animal/sample left out and each CV run output a pickle
file and prediction result, default 'train'
-c1 NUMLEVEL1, --numLevel1 NUMLEVEL1
integer, optional, number of classes at level 1,
number of experts = number of classes at level 1 x
number of classes at level 2, default 1
-c2 NUMLEVEL2, --numLevel2 NUMLEVEL2
integer, optional, number of classes at level 2,
default 5
-e EPOCHS, --epochs EPOCHS
integer, optional, number of epochs to train MESSI,
default 20
-gs GRID_SEARCH, --grid_search GRID_SEARCH
boolean, optional, if conduct grid search for hyper-
parameters, default False
-ns N_SETS, --n_sets N_SETS
integer, optional, number of CV sets for grid search,
default 3
-r NUMREPLICATES, --numReplicates NUMREPLICATES
integer, optional, number of times to run with same
set of parameters, default 1
-p PREPROCESS, --preprocess PREPROCESS
string, optional, the way to include neighborhood
information; neighbor_cat: include by concatenating
them to the cell own features; neighbor_sum: include
by addinding to the cell own features; anything
without 'neighbor': no neighborhood information will
be used as features; 'baseline': only baseline
features; default 'neighbor_cat'
-tr TOPKRESPONSES, --topKResponses TOPKRESPONSES
integer, optional, number of top dispersed responses
genes to model,default None (to include all response
genes)
-ts TOPKSIGNALS, --topKSignals TOPKSIGNALS
integer, optional, number of top dispersed signalling
genes to use as features, default None (to include all
signalling genes)See tutorials/MESSI for MERFISH hypothalamus, for a detailed intro on how to
- Train and test a MESSI model
- Analyze the model parameters to infer cell subtypes differ in signaling genes
- Train and test the data with other model configurations
We also prepared tutorials/results reprudction to reproduce MESSI's results shown in our manuscript.
- 2-20-2022:
-- Uploaded the jupyter notebook for reproducing MESSI's results shown in the manuscript
Check our paper published in Bioinformatics for more information. The pre-print version is available at biorxiv.
The software is an implementation of the method MESSI, jointly developed by Dongshunyi "Dora" Li, Jun Ding and Ziv Bar-Joseph from System Biology Group @ Carnegie Mellon University.
- dongshul at andrew.cmu.edu
This project is licensed under the MIT License - see the LICENSE file for details