treelite prediction 4x slower than xgboost
nikiforvln9 opened this issue · comments
Hi all,
I found treelite predictions are slower than xgboost. Is this normal? Code and models are like below.
import time
import numpy as np
import treelite
import treelite_runtime
import xgboost as xgb
def build_treelite():
model = treelite.Model.load('0.model', model_format='xgboost')
model.export_lib(toolchain='msvc', libpath='0.dll', compiler="failsafe", verbose=True)
def test_predicts():
data = np.array([np.random.uniform(0, 1, 792)])
# print(X)
X = xgb.DMatrix(data)
tree_model = xgb.Booster()
tree_model.load_model("0.model")
start = time.time()
y = tree_model.predict(X)
print('xgboost time: ', y, time.time() - start)
treelite_model = treelite_runtime.Predictor("0.dll")
X = treelite_runtime.DMatrix(data)
start = time.time()
y = treelite_model.predict(X)
print('treelite time: ', y, time.time() - start)
if __name__ == "__main__":
# build_treelite()
test_predicts()
Can you give a little more detail on the hardware you're running on? Are you using CPU XGBoost? What CPUs are available on your system?
One thing to be generally aware of is that the Treelite project is moving away from its previous work on code generation for model inference. It will continue to maintain GTIL as a reference implementation for evaluating Treelite trees, but its primary focus will be on offering a comprehensive model representation format that supports all the major tree model training frameworks.
Part of the reason for that is that cuML's Forest Inference Library (FIL) is now a dedicated project for providing accelerated inference for Treelite models on both CPU and GPU. The 23.04 release of cuML will include experimental support for CPU FIL and more complete support for any model that can be represented via Treelite. By letting FIL focus on the inference performance component, the Treelite project can focus more on its core value proposition: providing a comprehensive and unified exchange format for tree models.
If you're interested in trying FIL for yourself, read on, but otherwise feel free to ignore the rest, and we can try to give you more info on the behavior you're seeing based on your hardware details.
I took your script, modified it a bit, and used it to generate the following graph comparing available inference solutions for the model you provided:
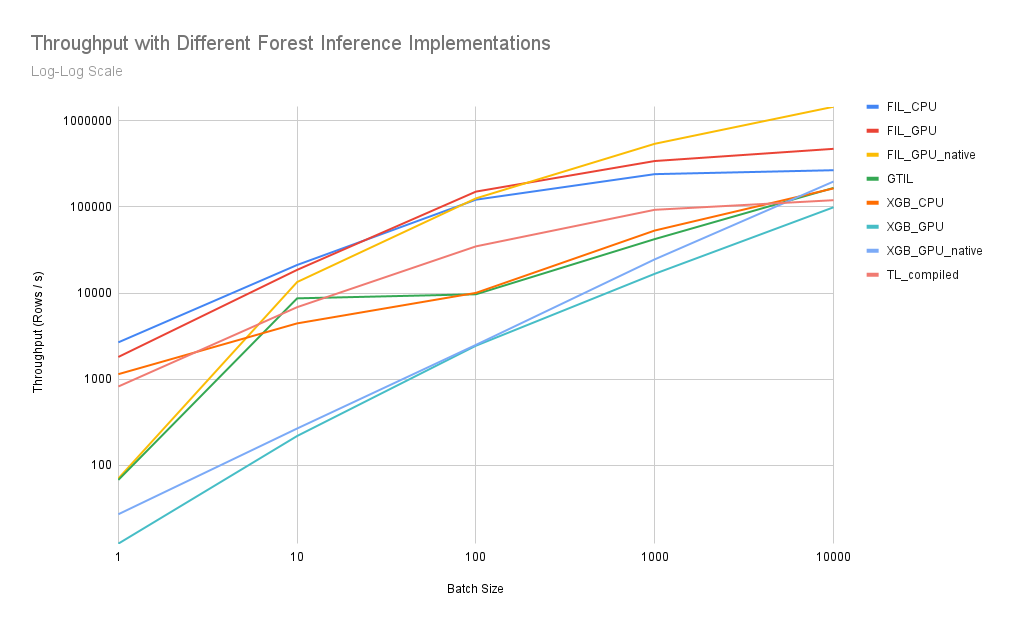
Your original script looked exclusively at batch size 1, but I've included larger batch sizes in case your actual application can take advantage of batched execution. You can see that at batch size 1, CPU FIL dominates, followed by GPU FIL. At higher batch sizes, GPU FIL unsurprisingly outperforms. The _native suffix for GPU execution methods indicates that input was provided on GPU (e.g via cupy arrays) as opposed to including the host->device transfer in the benchmark.
Hardware: GPU was a single NVIDIA A100 and CPU was 2x48 core Epyc 7642. The trends you see in these results may vary a bit depending on hardware and model details (especially model depth and number of trees).
If you want to experiment with FIL yourself, here is a simple example invocation using the unstable 23.04 nightly build:
import numpy as np
from cuml.common.device_selection import using_device_type
from cuml.experimental import ForestInference
model_path = '0.model'
X = np.random.uniform(0, 1, (1, 792))
with using_device_type('cpu'): # Or 'gpu' if you prefer
fil_model = ForestInference.load(model_path)
y = fil_model.predict(X)
For optimal performance, you can also tweak the chunk_size parameter in the predict call. If you are only focused on batch size 1, I'd just set it to 1 (fil_model.predict(X, chunk_size=1)). For larger batch sizes, you can set it to any power of 2 up to 32. In the benchmarking script I linked above, I added a quick search for the optimal chunk_size value, but you can also generally get reasonably good performance without optimizing that parameter.
Can you give a little more detail on the hardware you're running on? Are you using CPU XGBoost? What CPUs are available on your system?
One thing to be generally aware of is that the Treelite project is moving away from its previous work on code generation for model inference. It will continue to maintain GTIL as a reference implementation for evaluating Treelite trees, but its primary focus will be on offering a comprehensive model representation format that supports all the major tree model training frameworks.
Part of the reason for that is that cuML's Forest Inference Library (FIL) is now a dedicated project for providing accelerated inference for Treelite models on both CPU and GPU. The 23.04 release of cuML will include experimental support for CPU FIL and more complete support for any model that can be represented via Treelite. By letting FIL focus on the inference performance component, the Treelite project can focus more on its core value proposition: providing a comprehensive and unified exchange format for tree models.
If you're interested in trying FIL for yourself, read on, but otherwise feel free to ignore the rest, and we can try to give you more info on the behavior you're seeing based on your hardware details.
I took your script, modified it a bit, and used it to generate the following graph comparing available inference solutions for the model you provided:
Your original script looked exclusively at batch size 1, but I've included larger batch sizes in case your actual application can take advantage of batched execution. You can see that at batch size 1, CPU FIL dominates, followed by GPU FIL. At higher batch sizes, GPU FIL unsurprisingly outperforms. The
_nativesuffix for GPU execution methods indicates that input was provided on GPU (e.g via cupy arrays) as opposed to including the host->device transfer in the benchmark.Hardware: GPU was a single NVIDIA A100 and CPU was 2x48 core Epyc 7642. The trends you see in these results may vary a bit depending on hardware and model details (especially model depth and number of trees).
If you want to experiment with FIL yourself, here is a simple example invocation using the unstable 23.04 nightly build:
import numpy as np from cuml.common.device_selection import using_device_type from cuml.experimental import ForestInference model_path = '0.model' X = np.random.uniform(0, 1, (1, 792)) with using_device_type('cpu'): # Or 'gpu' if you prefer fil_model = ForestInference.load(model_path) y = fil_model.predict(X)For optimal performance, you can also tweak the
chunk_sizeparameter in the predict call. If you are only focused on batch size 1, I'd just set it to 1 (fil_model.predict(X, chunk_size=1)). For larger batch sizes, you can set it to any power of 2 up to 32. In the benchmarking script I linked above, I added a quick search for the optimalchunk_sizevalue, but you can also generally get reasonably good performance without optimizing that parameter.
Thanks! I'm working on an vps with 8-core CPU windows machine. Since cuML does not support building on windows, I cannot test the performance improvement.
Really thank you for your infomation about FIL! I will later try it when we port our project to linux.
Thanks! I'm working on an vps with 8-core CPU windows machine.
Ah! In that case, for batch size 1 with this model, I think a 4x differential is not out of the question. As you can see in the graph I posted, TL compiled models do a better job of exploiting larger batch sizes, but it starts out underperforming XGBoost CPU by 40%. On a slower machine, that differential could be magnified.
Since cuML does not support building on windows, I cannot test the performance improvement. Really thank you for your infomation about FIL! I will later try it when we port our project to linux.
We do support conda installs on WSL2 if you'd like to try it on Windows, but I realize that's not always a perfect solution. One of the use-cases we'd like to focus on in the future is exactly what you've described here: Prototyping/developing with cuML anywhere (regardless of hardware or OS) and then being able to move to a system with NVIDIA GPUs when you're ready to scale up. Hopefully we'll have more interesting developments in that space soon! Definitely don't want to leave any non-Linux folks behind if FIL is the primary solution for accelerated Treelite execution.