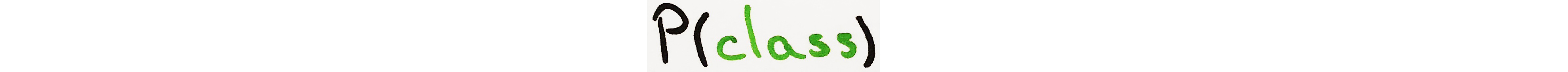Building a Naive Bayes classifier using Python.
The complete code for the below tutotial could be found in nb_tutorial.py
We will be using Naive Bayes and the Gaussian Distribution (Normal Distribution) to build a classifier in Python from scratch.
The Gauss Naive Bayes Classifier will be able to run on four classic data sets:
Here we'll be working with just the iris data set.
The logic for the code to work on all four data sets is in gauss_nb.py. This tutotial will follow the logic in nb_tutorial.py.
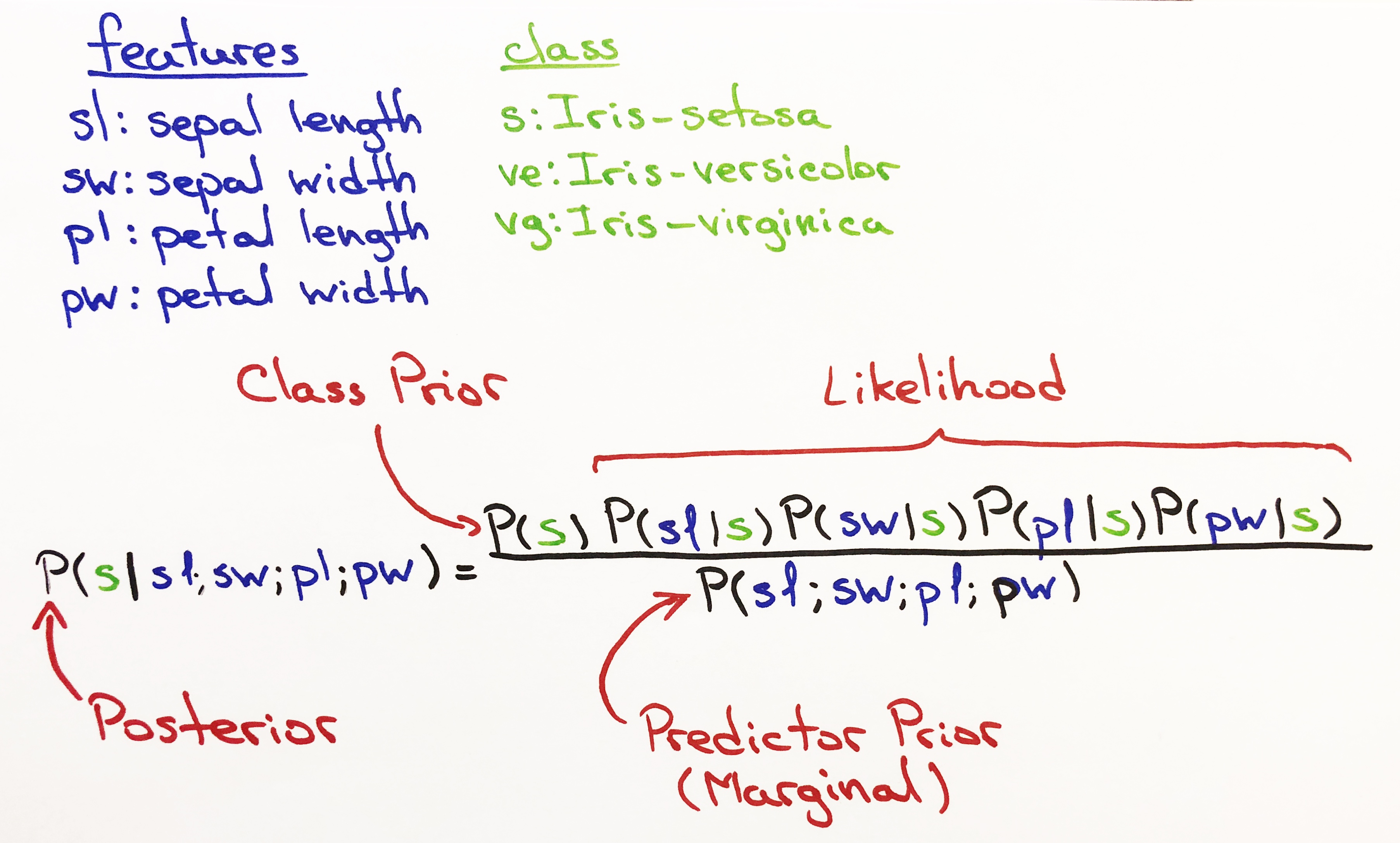
The Iris data set is a classic and is widely used when explaining classification models. The data set has 4 independent variables and 1 dependent variable that has 3 different classes.
The first 4 columns are the independent variables (features).
The 5th column is the dependent variable (class).
- sepal length (cm)
- sepal width (cm)
- petal length (cm)
- petal width (cm)
- classes:
- Iris Setosa,
- Iris Versicolour
- Iris Virginica
- 5 row sample from the Iris data
- The first 4 columns represent the features (sepal length, sepal width, petal length, petal width)
- The last column represents the class for each row. (Setosa, Versicolour, Virginica)
| sepal length | sepal width | petal length | petal width | class |
|---|---|---|---|---|
| 5.1 | 3.5 | 1.4 | 0.2 | Iris-setosa |
| 4.9 | 3.0 | 1.4 | 0.2 | Iris-setosa |
| 7.0 | 3.2 | 4.7 | 1.4 | Iris-versicolor |
| 6.3 | 2.8 | 5.1 | 1.5 | Iris-virginica |
| 6.4 | 3.2 | 4.5 | 1.5 | Iris-versicolor |
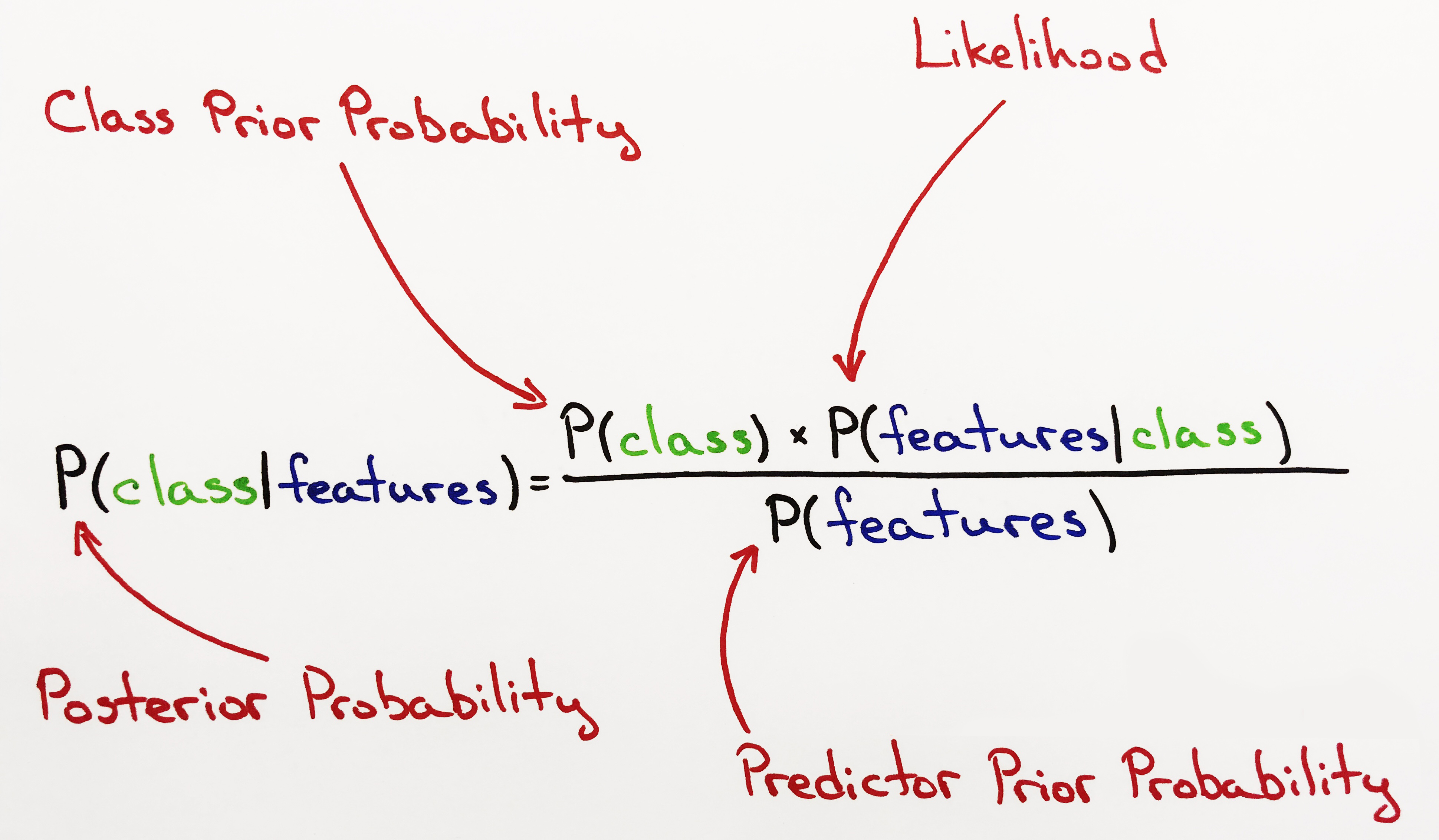
Posterior Probability:
- This is the updated belief given the new data.
Class Prior Probability:
- This is our Prior Belief. The probability of the class before updating our belief.
Likelihood:
- Likelihood is calculated by taking the product of all Normal Probability Density Functions also referred to as Gauss. Hence, the name Gauss Naive Bayes.
- We will use the Normal PDF formula to calculate the Normal Probability for each feature given the class. Likelihood is the product of all Normal PDFs.
- There's an important distinction to make between Likelihood and Probability. Normal Probability is calculated for each feature given the class and is always between 0 and 1. Likelihood is the product of all Normal Probabilites. The number of features is infinite and limited to our imagination. Hence, the product of all Normal Probabilities is not a probability but a Likelihood, because there will always be features that are not be accounted for.
Predictor Prior Probability:
- Predictor Prior Probability is the same as Marginal Probability. It is the probability given the new data under all possible features. It isn't necessary for a Naive Bayes Classifier to calculate this. The results do not change or change very little. Though we do calculate it here.
See Normal Distribution (Wikipedia) definition.
The Normal Distribution will help determine the Normal Probability for each new feature given the class. The product of all Normal Probabilities will result in the likelihood of a class occurring given the new features. In other words, for each column of our data set the Normal Distribution will calculate the Normal Probability and the product of all columns will equal the likelihood of that class occurring.
See Joint PDF (Wikipedia ) definition.
The Joint Probability is the product of all PDFs. In our case, the product of all Normal PDFs (Normal Probabilities). Multiplying all of the Normal Probabilities returns the likelihood.
Building the Naive Bayes Classifier.
Here, we'll create the structure and the methods to read and prepare data for modeling.
Every function is created from scratch. However, instead of having to download the data, we're using an api call to get the data.
$ pip install requests
Import the necessary libraries and create the class.
# -*- coding: utf-8 -*-
from collections import defaultdict
from math import pi
from math import e
import requests
import random
import csv
import re
class GaussNB:
def __init__(self):
pass
def main():
print "Here we will handle class methods."
if __name__ == '__main__':
main()Execute in terminal:
$ python nb_tutorial.py
Here we will handle class methods.
Read in the raw data and convert each string into an integer.
class GaussNB:
def __init__(self):
pass
def load_csv(self, data, header=False):
"""
:param data: raw comma seperated file
:param header: remove header if it exists
:return:
Load and convert each string of data into a float
"""
lines = csv.reader(data.splitlines())
dataset = list(lines)
if header:
# remove header
dataset = dataset[1:]
for i in range(len(dataset)):
dataset[i] = [float(x) if re.search('\d', x) else x for x in dataset[i]]
return dataset
def main():
nb = GaussNB()
url = 'http://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data'
data = requests.get(url).content
data = nb.load_csv(data, header=True)
print data[:3] # first 3 rows
if __name__ == '__main__':
main()[[4.9, 3.0, 1.4, 0.2, 'Iris-setosa'], [4.7, 3.2, 1.3, 0.2, 'Iris-setosa'], [4.6, 3.1, 1.5, 0.2, 'Iris-setosa']]
Split the data into a train set and a test set.
The weight will determine how much of the data will be in the train set.
class GaussNB:
.
.
.
def split_data(self, data, weight):
"""
:param data:
:param weight: indicates the percentage of rows that'll be used for training
:return:
Randomly selects rows for training according to the weight and uses the rest of the rows for testing.
"""
train_size = int(len(data) * weight)
train_set = []
for i in range(train_size):
index = random.randrange(len(data))
train_set.append(data[index])
data.pop(index)
return [train_set, data]
def main():
nb = GaussNB()
url = 'http://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data'
data = requests.get(url).content
data = nb.load_csv(data, header=True)
train_list, test_list = nb.split_data(data, weight=.67)
print "Using %s rows for training and %s rows for testing" % (len(train_list), len(test_list))
if __name__ == '__main__':
main()Group the data by class, by mapping each class to it's respective rows of data.
E.g. (This is a sample of what we mean using the table from above.)
{
'Iris-virginica': [
[6.3, 2.8, 5.1, 1.5],
], 'Iris-setosa': [
[5.1, 3.5, 1.4, 0.2],
[4.9, 3.0, 1.4, 0.2],
], 'Iris-versicolor': [
[7.0, 3.2, 4.7, 1.4],
[6.4, 3.2, 4.5, 1.5],
]
}class GaussNB:
.
.
.
def group_by_class(self, data, target):
"""
:param data: Training set. Lists of events (rows) in a list
:param target: Index for the target column. Usually the last index in the list
:return:
Mapping each target to a list of it's features
"""
target_map = defaultdict(list)
for index in range(len(data)):
features = data[index]
if not features:
continue
x = features[target]
target_map[x].append(features[:-1]) # designating the last column as the class column
print 'Identified %s different target classes: %s' % (len(target_map.keys()), target_map.keys())
return dict(target_map)
def main():
nb = GaussNB()
url = 'http://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data'
data = requests.get(url).content
data = nb.load_csv(data, header=True)
train_list, test_list = nb.split_data(data, weight=.67)
print "Using %s rows for training and %s rows for testing" % (len(train_list), len(test_list))
group = nb.group_by_class(data, -1)
print "Grouped into %s classes: %s" % (len(group.keys()), group.keys())
if __name__ == '__main__':
main()Grouped into 3 classes: ['Iris-virginica', 'Iris-setosa', 'Iris-versicolor']
Prepare the data for modeling. Mean and standard deviation is self explanatory. The summary is the combination of both and will be calculated for each feature.
Calculate the mean for [5.9, 3.0, 5.1, 1.8].
class GaussNB:
.
.
.
def mean(self, numbers):
"""
:param numbers: list of numbers
:return:
"""
result = sum(numbers) / float(len(numbers))
return result
def main():
nb = GaussNB()
print "Mean: %s" % nb.mean([5.9, 3.0, 5.1, 1.8])
if __name__ == '__main__':
main()Mean: 3.95
Calculate the standard deviation for [5.9, 3.0, 5.1, 1.8].
class GaussNB:
.
.
.
def stdev(self, numbers):
"""
:param numbers: list of numbers
:return:
Calculate the standard deviation for a list of numbers.
"""
avg = self.mean(numbers)
squared_diff_list = []
for num in numbers:
squared_diff = (num - avg) ** 2
squared_diff_list.append(squared_diff)
squared_diff_sum = sum(squared_diff_list)
sample_n = float(len(numbers) - 1)
var = squared_diff_sum / sample_n
return var ** .5
def main():
nb = GaussNB()
print "Standard Deviation: %s" % nb.stdev([5.9, 3.0, 5.1, 1.8])
if __name__ == '__main__':
main()Standard Deviation: 1.88414436814
Return the (mean, standard deviation) combination for each feature column of the data set.
class GaussNB:
.
.
.
def summarize(self, data):
"""
:param data: lists of events (rows) in a list
:return:
Use zip to line up each feature into a single column across multiple lists.
yield the mean and the stdev for each feature
"""
for attributes in zip(*data):
yield {
'stdev': self.stdev(attributes),
'mean': self.mean(attributes)
}
def main():
nb = GaussNB()
data = [
[5.9, 3.0, 5.1, 1.8],
[5.1, 3.5, 1.4, 0.2]
]
print "Feature Summary: %s" % [i for i in nb.summarize(data)]
if __name__ == '__main__':
main()Feature Summary:
[
{'mean': 5.5, 'stdev': 0.5656854249492386}, # sepal length
{'mean': 3.25, 'stdev': 0.3535533905932738}, # sepal width
{'mean': 3.25, 'stdev': 2.6162950903902256}, # petal length
{'mean': 1.0, 'stdev': 1.1313708498984762} # petal width
]
Building methods for calculating Bayes Theorem:
Prior Probability is what we know about each class before considering new information. It is simply the probability of each class occurring.
This method calculates the probability of each class.
class GaussNB:
.
.
.
def prior_prob(self, group, target, data):
"""
:return:
The probability of each target class
"""
total = float(len(data))
result = len(group[target]) / total
return result
def main():
nb = GaussNB()
url = 'http://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data'
data = requests.get(url).content
data = nb.load_csv(data, header=True)
train_list, test_list = nb.split_data(data, weight=.67)
print "Using %s rows for training and %s rows for testing" % (len(train_list), len(test_list))
group = nb.group_by_class(data, -1) # designating the last column as the class column
print "Grouped into %s classes: %s" % (len(group.keys()), group.keys())
for target_class in ['Iris-virginica', 'Iris-setosa', 'Iris-versicolor']:
prior_prob = nb.prior_prob(group, target_class, data)
print 'P(%s): %s' % (target_class, prior_prob)
if __name__ == '__main__':
main()Using 100 rows for training and 50 rows for testing
Grouped into 3 classes: ['Iris-virginica', 'Iris-setosa', 'Iris-versicolor']
P(Iris-virginica): 0.38
P(Iris-setosa): 0.3
P(Iris-versicolor): 0.32
Tying the previous methods together to determine the Prior Probability for each class and the (mean, standard deviation) combination for each feature of each class.
class GaussNB:
.
.
.
def train(self, train_list, target):
"""
:param data:
:param target: target class
:return:
For each target:
1. yield prior: the probability of each class. P(class) eg P(Iris-virginica)
2. yield summary: list of {'mean': 0.0, 'stdev': 0.0}
"""
group = self.group_by_class(train_list, target)
self.summaries = {}
for target, features in group.iteritems():
self.summaries[target] = {
'prior': self.prior_prob(group, target, train_list),
'summary': [i for i in self.summarize(features)],
}
return self.summaries
def main():
nb = GaussNB()
url = 'http://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data'
data = requests.get(url).content
data = nb.load_csv(data, header=True)
train_list, test_list = nb.split_data(data, weight=.67)
print "Using %s rows for training and %s rows for testing" % (len(train_list), len(test_list))
group = nb.group_by_class(data, -1) # designating the last column as the class column
print "Grouped into %s classes: %s" % (len(group.keys()), group.keys())
print nb.train(train_list, -1)
if __name__ == '__main__':
main()Using 100 rows for training and 50 rows for testing
Grouped into 3 classes: ['Iris-virginica', 'Iris-setosa', 'Iris-versicolor']
{'Iris-setosa': {'prior': 0.3,
'summary': [{'mean': 4.980000000000001, 'stdev': 0.34680810554104063}, # sepal length
{'mean': 3.406666666666667, 'stdev': 0.3016430104397023}, # sepal width
{'mean': 1.496666666666667, 'stdev': 0.20254132542705236}, # petal length
{'mean': 0.24333333333333343, 'stdev': 0.12228664272317624}]}, # petal width
'Iris-versicolor': {'prior': 0.31,
'summary': [{'mean': 5.96774193548387, 'stdev': 0.4430102307127106},
{'mean': 2.7903225806451615, 'stdev': 0.28560443356698495},
{'mean': 4.303225806451613, 'stdev': 0.41990782398659987},
{'mean': 1.3451612903225807, 'stdev': 0.17289439874755796}]},
'Iris-virginica': {'prior': 0.39,
'summary': [{'mean': 6.679487179487178, 'stdev': 0.585877428882027},
{'mean': 3.002564102564103, 'stdev': 0.34602036712733625},
{'mean': 5.643589743589742, 'stdev': 0.5215336048086158},
{'mean': 2.0487179487179477, 'stdev': 0.2927831916298213}]}}
We're looking to get calculate the Likelihood for each class. Likelihood is calculated by multiplying the Normal Probabilities of each feature.
For each feature given the class we calculate the Normal Probability using the Normal Distribution.
E.g.
As a quick example, below, we're using the Normal Distribution to calculate a single Normal Probability.
5 is the value of the sepal-width and {'mean': 4.98, 'stdev': 0.35} is the summary for Iris-setosa sepal-width.
We make this calculation for each feature and multiply all of the results together to get the Likelihood given that class.
Likelihood is a single value.
class GaussNB:
.
.
.
def normal_pdf(self, x, mean, stdev):
"""
:param x: a variable
:param mean: µ - the expected value or average from M samples
:param stdev: σ - standard deviation
:return: Gaussian (Normal) Density function.
N(x; µ, σ) = (1 / 2πσ) * (e ^ (x–µ)^2/-2σ^2
"""
variance = stdev ** 2
exp_squared_diff = (x - mean) ** 2
exp_power = -exp_squared_diff / (2 * variance)
exponent = e ** exp_power
denominator = ((2 * pi) ** .5) * stdev
pdf = exponent / denominator
return pdf
def main():
nb = GaussNB()
normal_pdf = nb.normal_pdf(5, 4.98, 0.35)
print normal_pdf
if __name__ == '__main__':
main()1.13797564994
 P(features) is referenced from Bayes Theorem.
P(features) is referenced from Bayes Theorem.
Below we break down Bayes Theorem further, and expand on calculating the Marginal Probability only using the Iris-setosa class. This is to be repeated for each class when running the prediction. The class with the highest posterior probability is the predicted class.
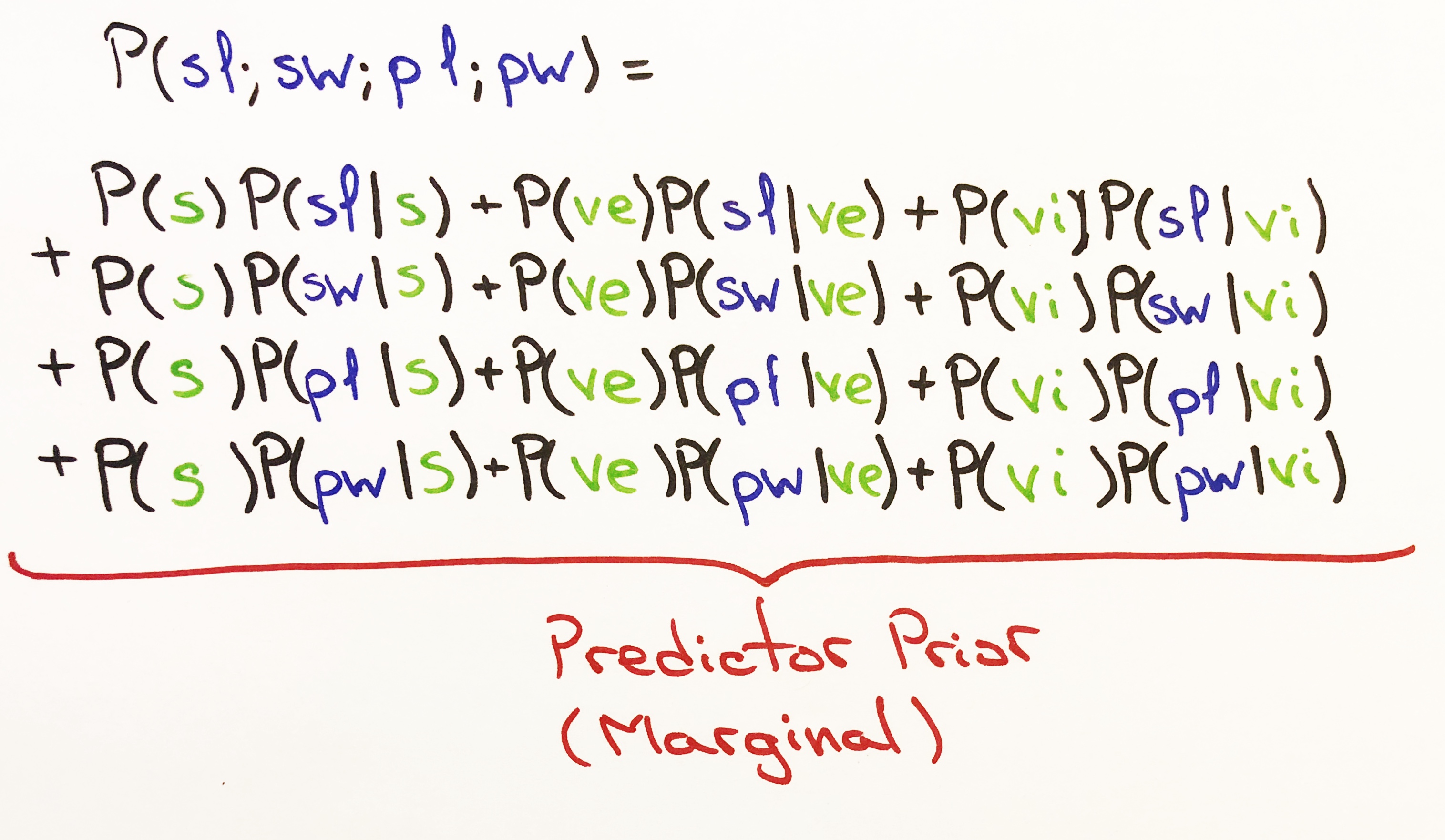
The marginal probability is calculated using the sum of the product of the Prior Probability and Normal Probability.
The Marginal Probability is determined using all 3 classes and the Normal Probability of their features. The Marginal value, a single value, will be the same across all classes for each test. We could think of the Marginal Probability as the total probability of all 3 classes occurring given the Normal Probability of each class. Thus, the Marginal value will be the same across all classes.
Reminder, to predict the class, we're looking for the highest Posterior Probability from among all possible classes. Dividing by the same value will not improve the accuracy of predicting the correct class.
For the purposes of sticking to the true Bayes Theorem, we'll use it here.
class GaussNB:
.
.
.
def marginal_pdf(self, pdfs):
"""
:param pdfs: list of probability densities for each feature
:return:
Marginal Probability Density Function (Predictor Prior Probability)
Summing up the product of P(class) prior probability and the probability density of each feature P(feature | class)
marginal pdf =
P(setosa) * P(sepal length | setosa) + P(versicolour) * P(sepal length | versicolour) + P(virginica) * P(sepal length | virginica)
+ P(setosa) * P(sepal width | setosa) + P(versicolour) * P(sepal width | versicolour) + P(virginica) * P(sepal width | virginica)
+ P(setosa) * P(petal length | setosa) + P(versicolour) * P(petal length | versicolour) + P(virginica) * P(petal length | virginica)
+ P(setosa) * P(petal width | setosa) + P(versicolour) * P(petal width | versicolour) + P(virginica) * P(petal width | virginica)
"""
predictors = []
for target, features in self.summaries.iteritems():
prior = features['pr ior']
for index in range(len(pdfs)):
normal_pdf = pdfs[index]
predictors.append(prior * normal_pdf)
marginal_pdf = sum(predictors)
return marginal_pdf
def main():
nb = GaussNB()
url = 'http://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data'
data = requests.get(url).content
data = nb.load_csv(data, header=True)
train_list, test_list = nb.split_data(data, weight=.67)
print "Using %s rows for training and %s rows for testing" % (len(train_list), len(test_list))
group = nb.group_by_class(data, -1) # designating the last column as the class column
print "Grouped into %s classes: %s" % (len(group.keys()), group.keys())
nb.train(train_list, -1)
marginal_pdf = nb.marginal_pdf(pdfs=[0.02, 0.37, 3.44e-12, 4.35e-09])
print marginal_pdf
if __name__ == '__main__':
main()Using 100 rows for training and 50 rows for testing
Grouped into 3 classes: ['Iris-virginica', 'Iris-setosa', 'Iris-versicolor']
0.38610000431
Posterior Probability is our update believe given new features (data). We have to calculate the Posterior Probability for each class. The predicted class is the class with the highest Posterior Probability.
This method takes a single test_row and returns the Posterior Probability for each class.
Using Bayes Theorem from above:
class GaussNB:
.
.
.
def posterior_probabilities(self, test_row):
"""
:param test_row: single list of features to test
:return:
For each feature (x) in the test_row:
1. Calculate Predictor Prior Probability using the Normal PDF N(x; µ, σ). eg = P(feature | class)
2. Calculate Likelihood by getting the product of the prior and the Normal PDFs
3. Multiply Likelihood by the prior to calculate the Joint PDF. P(Iris-virginica)
E.g.
prior: P(setosa)
likelihood: P(sepal length | setosa) * P(sepal width | setosa) * P(petal length | setosa) * P(petal width | setosa)
numerator (joint pdf): prior * likelihood
denominator (marginal pdf): predictor prior probability
posterior_prob = joint pdf/ marginal pdf
returning a dictionary mapping of class to it's posterior probability
"""
posterior_probs = {}
for target, features in self.summaries.iteritems():
total_features = len(features['summary'])
likelihood = 0
pdfs = []
for index in range(total_features):
mean = features['summary'][index]['mean']
stdev = features['summary'][index]['stdev']
x = test_row[index]
normal = self.normal_pdf(x, mean, stdev)
likelihood = posterior_probs.get(target, 1) * normal
pdfs.append(normal)
marginal = self.marginal_pdf(pdfs)
prior = features['prior']
posterior_probs[target] = (prior * likelihood) / marginal
return posterior_probs
def main():
nb = GaussNB()
url = 'http://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data'
data = requests.get(url).content
data = nb.load_csv(data, header=True)
train_list, test_list = nb.split_data(data, weight=.67)
print "Using %s rows for training and %s rows for testing" % (len(train_list), len(test_list))
group = nb.group_by_class(data, -1) # designating the last column as the class column
print "Grouped into %s classes: %s" % (len(group.keys()), group.keys())
nb.train(train_list, -1)
posterior_probs = nb.posterior_probabilities([6.3, 2.8, 5.1, 1.5]) # 'Iris-virginica'
print "Posterior Probabilityies: %s" % posterior_probs
if __name__ == '__main__':
main()Using 100 rows for training and 50 rows for testing
Grouped into 3 classes: ['Iris-virginica', 'Iris-setosa', 'Iris-versicolor']
Posterior Probabilityies: {
'Iris-virginica': 0.01402569010123345,
'Iris-setosa': 2.6269216431943473e-26,
'Iris-versicolor': 0.14165560618269524
}
This method will call our posterior_probabilities() method on a single test vector eg ([6.3, 2.8, 5.1, 1.5]).
The test vector is a list of features. For each test vector we will calculate 3 Posterior Probabilities; one for each class.
The Posterior Probability is the updated believe given the new data, test vector.
The get_prediction() method will simply choose the highest Posterior Probability and return the predicted class for the given test_row.
class GaussNB:
.
.
.
def get_prediction(self, test_row):
"""
:param test_row: single list of features to test
:return:
Return the target class with the largest/best posterior probability
"""
posterior_probs = self.posterior_probabilities(test_row)
best_target = max(posterior_probs, key=posterior_probs.get)
return best_target
def main():
nb = GaussNB()
url = 'http://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data'
data = requests.get(url).content
data = nb.load_csv(data, header=True)
train_list, test_list = nb.split_data(data, weight=.67)
print "Using %s rows for training and %s rows for testing" % (len(train_list), len(test_list))
group = nb.group_by_class(data, -1) # designating the last column as the class column
print "Grouped into %s classes: %s" % (len(group.keys()), group.keys())
nb.train(train_list, -1)
prediction = nb.get_prediction([6.3, 2.8, 5.1, 1.5]) # 'Iris-virginica'
print 'According to the test row the best prediction is: %s' % prediction
if __name__ == '__main__':
main()Using 100 rows for training and 50 rows for testing
Grouped into 3 classes: ['Iris-virginica', 'Iris-setosa', 'Iris-versicolor']
According to the test row the best prediction is: Iris-versicolor
This method will return a prediction for each list (row).
Example input, list of lists:
[
[5.1, 3.5, 1.4, 0.2],
[4.9, 3.0, 1.4, 0.2],
]
For testing this method, we'll use the data from the sample data above.
class GaussNB:
.
.
.
def predict(self, test_set):
"""
:param test_set: list of features to test on
:return:
Predict the likeliest target for each row of the test_set.
Return a list of predicted targets.
"""
predictions = []
for row in test_set:
result = self.get_prediction(row)
predictions.append(result)
return predictions
def main():
nb = GaussNB()
url = 'http://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data'
data = requests.get(url).content
data = nb.load_csv(data, header=True)
train_list, test_list = nb.split_data(data, weight=.67)
print "Using %s rows for training and %s rows for testing" % (len(train_list), len(test_list))
group = nb.group_by_class(data, -1) # designating the last column as the class column
print "Grouped into %s classes: %s" % (len(group.keys()), group.keys())
nb.train(train_list, -1)
test = {
'Iris-virginica': [
[6.3, 2.8, 5.1, 1.5],
], 'Iris-setosa': [
[5.1, 3.5, 1.4, 0.2],
[4.9, 3.0, 1.4, 0.2],
], 'Iris-versicolor': [
[7.0, 3.2, 4.7, 1.4],
[6.4, 3.2, 4.5, 1.5],
]
}
for target, features in test.iteritems():
predicted = nb.predict(features)
print 'predicted target: %s | true target: %s' % (predicted, target)
if __name__ == '__main__':
main()Using 100 rows for training and 50 rows for testing
Grouped into 3 classes: ['Iris-virginica', 'Iris-setosa', 'Iris-versicolor']
predicted target: ['Iris-versicolor'] | true target: Iris-virginica
predicted target: ['Iris-setosa', 'Iris-setosa'] | true target: Iris-setosa # both test rows were predicted to be setosa
predicted target: ['Iris-versicolor', 'Iris-versicolor'] | true target: Iris-versicolor # both test rows were predicted to be versicolor
Accuracy will test the performance of the model by taking the total of correct predictions and dividing them by the total of predictions.
class GaussNB:
.
.
.
def accuracy(self, test_set, predicted):
"""
:param test_set: list of test_data
:param predicted: list of predicted classes
:return:
Calculate the the average performance of the classifier.
"""
correct = 0
actual = [item[-1] for item in test_set]
for x, y in zip(actual, predicted):
if x == y:
correct += 1
return correct / float(len(test_set))
def main():
nb = GaussNB()
url = 'http://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data'
data = requests.get(url).content
data = nb.load_csv(data, header=True)
train_list, test_list = nb.split_data(data, weight=.67)
print "Using %s rows for training and %s rows for testing" % (len(train_list), len(test_list))
group = nb.group_by_class(data, -1) # designating the last column as the class column
print "Grouped into %s classes: %s" % (len(group.keys()), group.keys())
nb.train(train_list, -1)
predicted = nb.predict(test_list)
accuracy = nb.accuracy(test_list, predicted)
print 'Accuracy: %.3f' % accuracy
if __name__ == '__main__':
main()Using 100 rows for training and 50 rows for testing
Grouped into 3 classes: ['Iris-virginica', 'Iris-setosa', 'Iris-versicolor']
Accuracy: 0.960
The above code will only work with the Iris Data set. You could find the logic in nb_tutorial.py
- Oleh Dubno - github.odubno
- Danny Argov
See also the list of contributors who participated in this project.
A hat tip to the authors that made this tutorial possible:
| Author | URL |
|---|---|
| Dr. Jason Brownlee | How To Implement Naive Bayes From Scratch in Python |
| Chris Albon | Naive Bayes Classifier From Scratch |
| Sunil Ray | 6 Easy Steps to Learn Naive Bayes Algorithm |
| Rahul Saxena | How The Naive Bayes Classifier Works In Machine Learning |
| Data Source | UCI Machine Learning |
| C. Randy Gallistel | Bayes for Beginners: Probability and Likelihood |
Project for Columbia Probability and Statistics course - Prof. Banu Baydil