Deep Negative Volume Segmentation
This is the official repository of the paper entitled "Deep negative volume segmentation", Nature Scientific Reports 11, 16292 (2021). https://doi.org/10.1038/s41598-021-95526-1
Clinical examination of three-dimensional image data of compound anatomical objects, such as complex joints, remains a tedious process, demanding the time and the expertise of physicians. For instance, automation of the segmentation task of the TMJ (temporomandibular joint) has been hindered by its compound three-dimensional shape, multiple overlaid textures, an abundance of surrounding irregularities in the skull, and a virtually omnidirectional range of the jaw’s motion—all of which extend the manual annotation process to more than an hour per patient. To address the challenge, we invent a new workflow for the 3D segmentation task: namely, we propose to segment empty spaces between all the tissues surrounding the object—the so-called negative volume segmentation. Our approach is an end-to-end pipeline that comprises a V-Net for bone segmentation, a 3D volume construction by inflation of the reconstructed bone head in all directions along the normal vector to its mesh faces. Eventually confined within the skull bones, the inflated surface occupies the entire “negative” space in the joint, effectively providing a geometrical/topological metric of the joint’s health. We validate the idea on the CT scans in a 50-patient dataset, annotated by experts in maxillofacial medicine, quantitatively compare the asymmetry given the left and the right negative volumes, and automate the entire framework for clinical adoption.
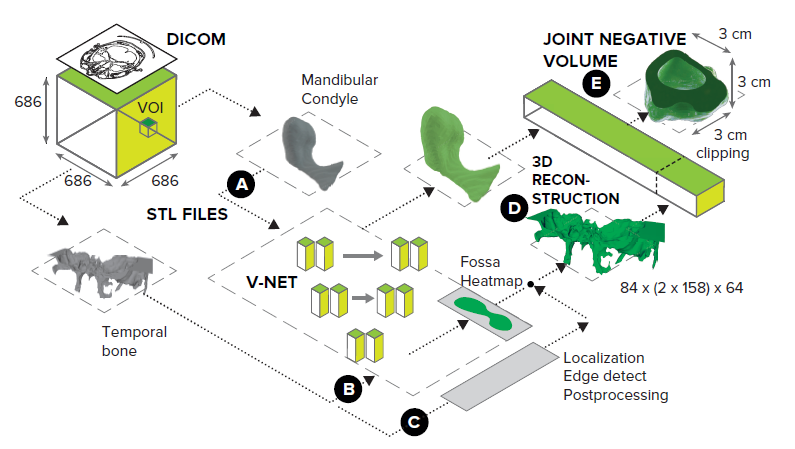
Fig. 1. End-to-end pipeline for Deep Negative Volume Segmentation. As an example, we take the most complex object in a human body - temporomandibular joint (TMJ), consisting of the mandibular condyle (MC) and the temporal bone (TB). Segmentation of MC and TB are shown as step A and step B, respectively. Step C and step D represent classical image enhancement of TB and 3D reconstruction of both bones. The “inflation/clipping” block represented by Step E.
Requirements
To install requirements:
pip install -r requirements.txt
Training
To train the models used in the paper, run this command:
python train.py --config <path_to_config_file>
where path_to_config_file is the path to a configuration file, which specifies all aspects of the training procedure. See e.g. config.txt for example how to specify training a standard V-Net with Dice + Cross-Entropy loss.
Evaluation
To evaluate models, run:
python eval.py --config <path_to_config_file>
Pre-trained Models
You can download pretrained models here:
- Pre-trained V-Net model for object loclization: https://drive.google.com/drive/folders/1qUtlMfNEBMQakJpmGQWiKTrwY59vpbom?usp=sharing
- 3D U-Net and V-Net for spherical negative volumes segmentation: https://drive.google.com/drive/folders/1-Ctq56kAMF3B24SdLJ_i00zc5Rb1rIze?usp=sharing
- 3D U-Net, 3D U-Net with attention gates, and V-Net trained with different loss function on mandibular condyle: https://drive.google.com/drive/folders/1uODu_VFmaOmVgWD7GNQ0kjdnZtQRDe7c?usp=sharing
- 3D U-Net, 3D U-Net with attention gates, and V-Net trained with different loss function on temporal bone: https://drive.google.com/drive/folders/1hLwS0J09u6Qz5_cpz4a-MyKEmnjeGVh4?usp=sharing
Results
Table 1. Mandibular condyle (MC), temporal bone (TB) and negative volume (NV) segmentation results. Notice that the whole-object 3D segmentation of the manually annotated “balls” from Fig.1 need more data to work properly, justifying the development of our automated pipeline which just needs MC and TB masks.| Obj. | Score | 3D U-Net | 3D U-Net+Att. | V-Net CE | V-Net D | V-Net D+CE |
|---|---|---|---|---|---|---|
| MC | DICE | 91.4 ± 5.3 | 89.8 ± 8.2 | 90.9 ± 4.5 | 90.9 ± 6.3 | 91.4 ± 4.8 |
| CE | 0.320 ± 0.003 | 0.320 ± 0.005 | 0.201 ± 0.075 | 0.175 ± 0.024 | 0.154 ± 0.053 | |
| HD | 14.7 ± 20.8 | 15.2 ± 21.6 | 11.9 ± 15.7 | 11.5 ± 20.1 | 10.5 ± 21.2 | |
| TB | DICE | 75.5 ± 8.8 | 75.8 ± 8.4 | 75.9 ± 6.9 | 76.7 ± 6.8 | 76.3 ± 7.2 |
| CE | 0.463 ± 0.043 | 0.462 ± 0.035 | 0.383 ± 0.088 | 0.396 ± 0.093 | 0.416 ± 0.100 | |
| HD | 29.8 ± 11.5 | 29.9 ± 11.3 | 27.9 ± 11.5 | 28.3 ± 10.7 | 27.6 ± 10.9 | |
| NV | DICE | 78.0 ± 10.6 | 77.8 ± 9.6 | 78.1 ± 8.8 | 78.2 ± 8.3 | 77.7 ± 7.7 |
| CE | 0.344 ± 0.016 | 0.349 ± 0.012 | 0.402 ± 0.019 | 0.396 ± 0.024 | 0.406 ± 0.022 | |
| HD | 15.8 ± 18.8 | 15.5 ± 17.6 | 19.1 ± 16.2 | 18.3 ± 16.9 | 18.7 ± 17.8 |
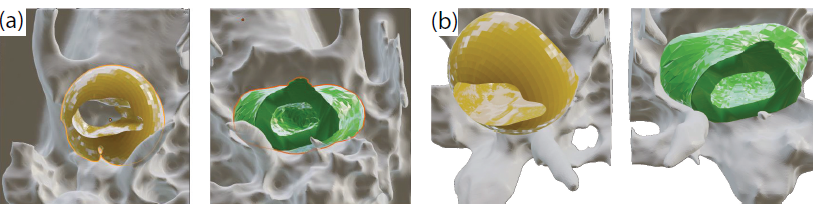
Fig. 2. Rendered regions of the TB (gray) featuring manually annotated negative volume (yellow), and a machine-generated one (green). Views: (a) axial, from bottom (b) same, tilted.
Citing
If you use this package in your publications or in other work, please cite it as follows:
@Article{Belikova2021,
author = {Kristina Belikova and Oleg Y. Rogov and Aleksandr Rybakov and Maxim V. Maslov and Dmitry V. Dylov},
title = {Deep negative volume segmentation},
journal = {Scientific Reports},
year = {2021},
volume = {11},
number = {1},
month = {aug},
doi = {10.1038/s41598-021-95526-1},
publisher = {Springer Science and Business Media {LLC}},
}
Maintainers
Kristina Belikova (Main contributor) @krisbell
Oleg Rogov @olegrgv