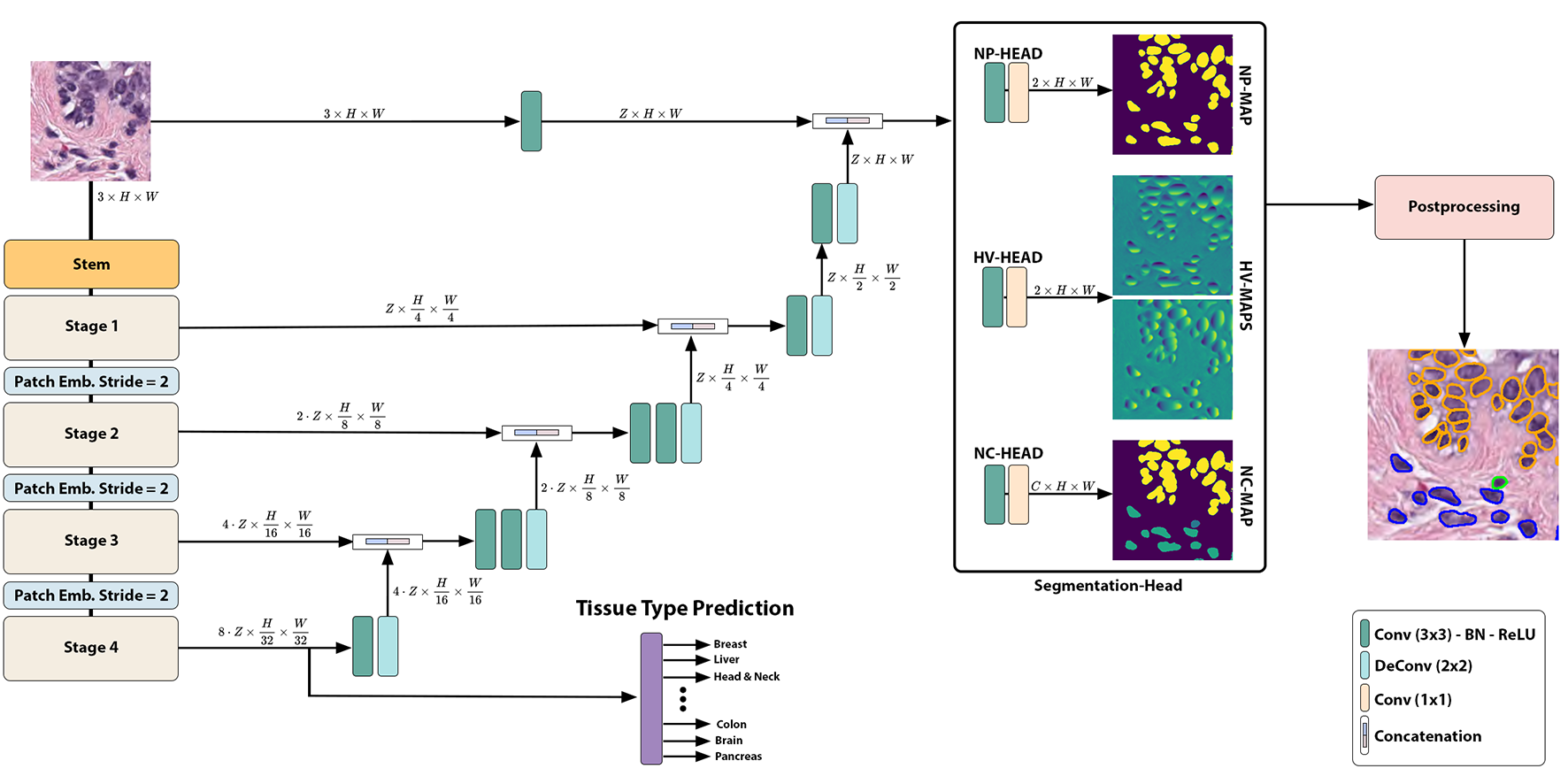
This repository contains the original PyTorch implementation of NuLite.
Tommasino C., Russo C., Rianldi A.M. (2024). NuLite - Lightweight and Fast Model for Nuclei Instance Segmentation and Classification. https://doi.org/10.48550/arXiv.2408.01797
- State-of-the-Art Performance for lightweight model for nuclei instance segmentation and classification;
- The project uses FastViT;
- U-Net Architecture: NuLite is a U-Net-like encoder-decoder network;
- Fast Inference on Gigapixel WSI: the framework provides fast inference results by utilizing a large inference patch size of 1024X1024 pixels. This approach enables efficient analysis of Gigapixel Whole Slide Images (WSI) and generates localizable deep features that hold potential value for downstream tasks. We provide a fast inference pipeline with connection to current Viewing Software such as QuPath (inherited from CellVIT).
Features • Code Notes • Repository Structure • Datasets • Set Up Environment • Training • Inference • Inference Examples • Authors • Citation
- Training
- Tile Inference
- WSI Inference
The training and inference code are taken from CellVIT repository.
The goal our this repository as well as the paper is to provide a lightweight model for nuclei instance segmentation and classification.
Below are the main directories in the repository:
├── base_ml # Foundational machine learning utilities and base classes
├── models # Contains model definitions and architectures
│ ├── encoders # FastViT implementation as encoder
├── nuclei_detection # Nuclei detection-specific modules and scripts
│ ├── datamodel # Data models or schemas for handling nuclei detection data
│ ├── datasets # Datasets and data loaders for nuclei detection
│ ├── experiments # Experiment scripts
│ ├── inference # Scripts for model inference and predictions
│ ├── training # Training scripts and configurations for nuclei detection models
│ ├── utils # Utility functions for nuclei detection
├── utils # General utilities and helper functions used across the project
├── preprocessing # Modules for preprocessing data before model inference
│ ├── encoding # Data encoding and transformation modules
│ │ ├── datasets # Encoded datasets or scripts handling the encoded data
│ └── patch_extraction # Logic and scripts for extracting patches from WSI
│ ├── src # Source code for patch extraction
│ │ ├── data # Raw or processed data related to patch extraction
│ │ │ └── tissue_detector.pt # Model file for detecting tissue areas
│ │ ├── utils # Utilities specific to patch extraction
├── README.md # Project overview and usage instructions
├── LICENSE # Legal terms for project usage and distributionBelow are the main executable scripts in the repository:
process_pannuke_dataset.py: script to transform PanNuke dataset for knowledge distillationtrain_nulite.py: main training script for NuLiterun_inference_wsi.py: main inference script for WSIpreprocessing/patch_extraction/main_extraction.py: main script for patches extraction for WSI
To run this code set up python environment as follows:
git clone https://github.com/DIAGNijmegen/NuLite.git
cd NuLite
python -m venv NuLite
source NuLite/bin/activate
pip install -r requirements.txtWe tested our code with Python 3.8.10/3.10.8 and Cuda 12.
Note
We used the training workflow provided by CellViT removing the metrics computation during the training step, for any further information see here.
To train the NuLite Network, use the train_nulite.py script from the command line interface (CLI). Below are the usage details and configuration requirements for running the script:
usage: train_nulite.py [-h] --config CONFIG [--gpu GPU] | --checkpoint CHECKPOINT]This script initiates an experiment using the specified configuration file.
-h, --help: Display the help message and exit.--gpu GPU: Specify the CUDA-GPU ID to use (default is None).--checkpoint CHECKPOINT: Provide the path to a PyTorch checkpoint file. The training will resume from this checkpoint with the settings provided. Note that this parameter cannot be set within the configuration file (default is None).
--config CONFIG: Specify the path to the configuration file (default is None). An example configuration file is available atconfigs_examples/traning_example.yaml. We used the same dataset structure of CellViT for the PanNuke, for more detail see here. The preparation script is in thenuclei_detection/datasets/folder.
Note
We used the inference provided by CellViT, for any further information see here.
Model checkpoints are available for download here:
License: Apache 2.0 with Commons Clause
The provided checkpoints were trained on whole PanNuke dataset.
To perform preprocessing, follow these steps:
- Prepare WSI using our preprocessing pipeline.
- Run inference with the
inference/cell_detection.pyscript.
To perform inference, use the cell_detection.py script in the cell_segmentation/inference folder:
usage: run_wsi_inference.py --model MODEL [--gpu GPU] [--magnification MAGNIFICATION] [--mixed_precision]
[--batch_size BATCH_SIZE] [--outdir_subdir OUTDIR_SUBDIR]
[--geojson] {process_wsi,process_dataset} ...
Perform NuLite inference for given run-directory with model checkpoints and logs.
optional arguments:
-h, --help show this help message and exit
--gpu GPU CUDA-GPU ID for inference. Default: 0 (default: 0)
--magnification MAGNIFICATION
Network magnification. Ensures correct resolution for the network. Default: 40 (default: 40)
--mixed_precision Use mixed precision for inference. Default: False (default: False)
--batch_size BATCH_SIZE
Inference batch size. Default: 8 (default: 8)
--outdir_subdir OUTDIR_SUBDIR
Creates a subdirectory in the cell_detection folder for storing results. Default: None (default: None)
--geojson Exports results as geojson files for use in software like QuPath. Default: False (default: False)
required named arguments:
--model MODEL Model checkpoint file for inference (default: None)
subcommands:
Main command for performing inference on a single WSI file or an entire dataset.
{process_wsi,process_dataset}To process a single WSI file, use the process_wsi subcommand:
usage: cell_detection.py process_wsi --wsi_path WSI_PATH --patched_slide_path PATCHED_SLIDE_PATH
Process a single WSI file.
arguments:
-h, --help show this help message and exit
--wsi_path WSI_PATH Path to WSI file.
--patched_slide_path PATCHED_SLIDE_PATH
Path to patched WSI file (specific WSI file, not parent path of patched slide dataset).To process an entire dataset, use the process_dataset subcommand:
usage: cell_detection.py process_dataset --wsi_paths WSI_PATHS --patch_dataset_path PATCH_DATASET_PATH [--filelist FILELIST] [--wsi_extension WSI_EXTENSION]
Process a whole dataset.
arguments:
-h, --help show this help message and exit
--wsi_paths WSI_PATHS
Path to the folder containing all WSI.
--patch_dataset_path PATCH_DATASET_PATH
Path to the folder containing the patch dataset.
--filelist FILELIST CSV file listing WSI to process. Each row should denote a filename (column named 'Filename'). If not provided, all WSI files with the given extension in the WSI folder are processed. Default: 'None'.
--wsi_extension WSI_EXTENSION
The file extension for the WSI files, as defined in configs.python.config (WSI_EXT). Default: 'svs'.We provide an example TCGA file to show the performance and usage of our algorithms. Files and scripts can be found in the example folder. The TCGA slide must be downloaded here: https://portal.gdc.cancer.gov/files/f9147f06-2902-4a64-b293-5dbf9217c668. Please place this file in the example folder.
python3 ./preprocessing/patch_extraction/main_extraction.py --config ./example/preprocessing_example.yamlOutput is stored inside ./example/output/preprocessing
Inference:
Download the models and store them in ./models/pretrained or on your preferred location and change the model parameter.
python3 ./cell_segmentation/inference/cell_detection.py \
--model ./models/pretrained/NuLite/NuLite-H-Weights.pth\
--gpu 0 \
--geojson \
process_wsi \
--wsi_path ./example/TCGA-V5-A7RE-11A-01-TS1.57401526-EF9E-49AC-8FF6-B4F9652311CE.svs \
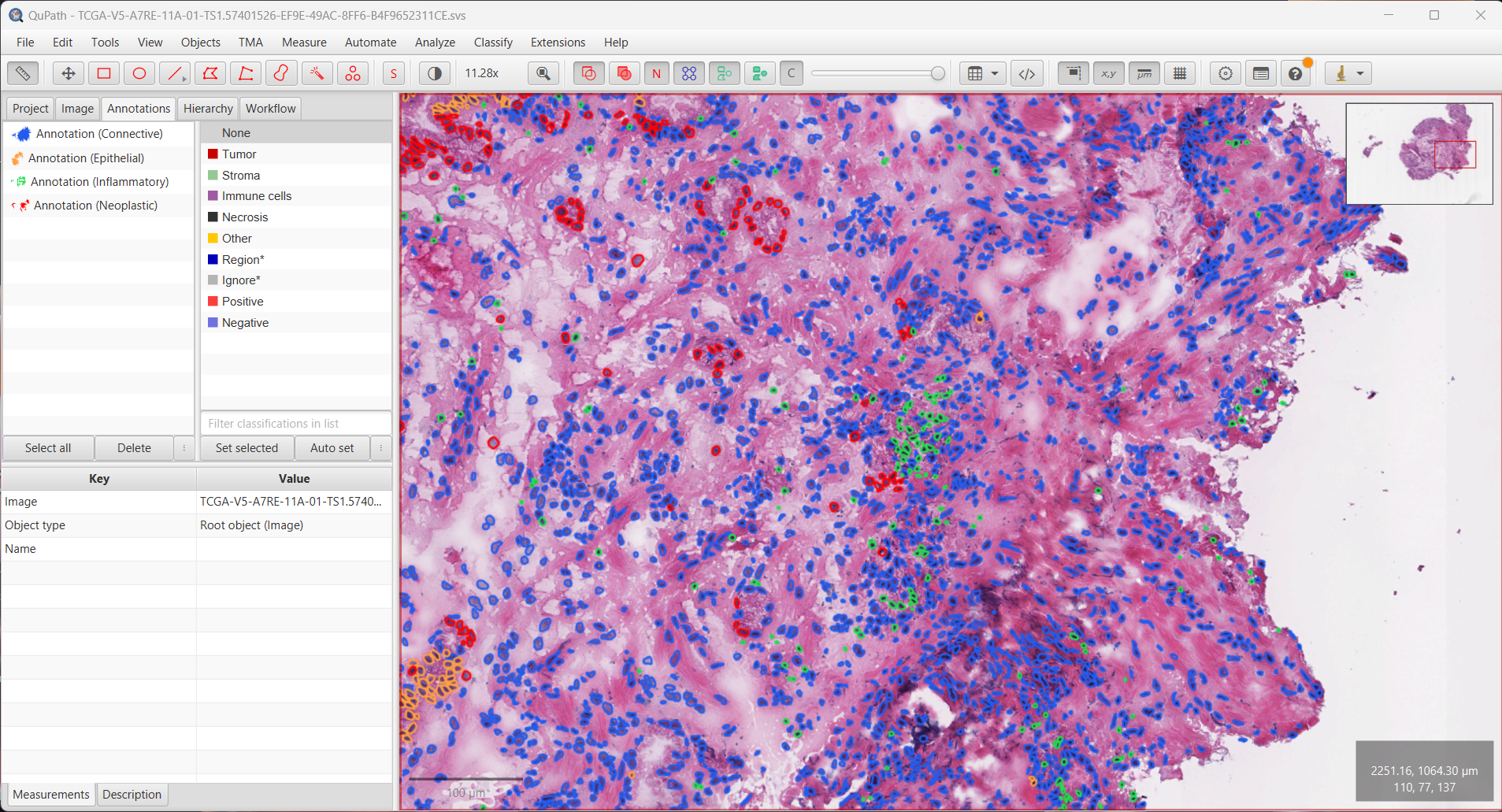
--patched_slide_path ./example/output/preprocessing/TCGA-V5-A7RE-11A-01-TS1.57401526-EF9E-49AC-8FF6-B4F9652311CEYou can import your results (.geojson files) into QuPath. The results should look like this:
@article{tommasino2024nulite,
title={NuLite - Lightweight and Fast Model for Nuclei Instance Segmentation and Classification},
author={Tommasino, Cristian and Russo, Cristiano and Rinaldi, Antonio Maria},
journal={arXiv preprint arXiv:2408.01797},
year={2024}
}
[1] Gamper, J., Alemi Koohbanani, N., Benet, K., Khuram, A., & Rajpoot, N. (2019). PanNuke: an open pan-cancer histology dataset for nuclei instance segmentation and classification. In Digital Pathology: 15th European Congress, ECDP 2019, Warwick, UK, April 10–13, 2019, Proceedings 15 (pp. 11-19). Springer International Publishing.
[2] Graham, S., Vu, Q. D., Raza, S. E. A., Azam, A., Tsang, Y. W., Kwak, J. T., & Rajpoot, N. (2019). Hover-net: Simultaneous segmentation and classification of nuclei in multi-tissue histology images. Medical image analysis, 58, 101563.
[2] Hörst, F., Rempe, M., Heine, L., Seibold, C., Keyl, J., Baldini, G., ... & Kleesiek, J. (2024). Cellvit: Vision transformers for precise cell segmentation and classification. Medical Image Analysis, 94, 103143.
This project is licensed under the APACHE License 2.0 - see the LICENSE file for details.
Note that the PanNuke dataset is licensed under Attribution-NonCommercial-ShareAlike 4.0 International, therefore the derived weights for NuLite are also shared under the same license. Please consider the implications of using the weights under this license on your work, and it's licensing.