Please refer to the User Manual
Update: I-VIP.2021
https://github.com/caozhichongchong/I-VIP/releases
For python >= 3.0
https://github.com/caozhichongchong/I-VIP/releases/download/2021/I-VIP.2021release.zip
For python < 3.0
https://github.com/caozhichongchong/I-VIP/releases/download/2021/I-VIP.py3.2021release.zip
unzip I-VIP.2020release.zip
-
Input folder (-i example), input files in the format of .gbff.gz (-f .gbff.gz), use GbffParser.py in scripts to extract contigs and ORFs (--ot 1), provide genbank files (--g example), use Module A2 (local search, --m 2), annotate the ORFs of antibiotic and metal resistance (--a Y), use usearch (--u usearch), output folder (example/example_output), input taxonomy metadata (--tx taxon.txt) and columns for phylum to strain level (--tc 4,10)
python I-VIP.py -i example -f .gbff.gz --ot 1 --g example --a Y --m 2 --t 1 --u usearch --r example/example_output --tc 4,10 --tx taxon.txt -
Input folder (-i example), input files in the format of .gbff (-f .gbff), use GbffParser.py in scripts to extract contigs and ORFs (--ot 1), provide genbank files (--g example), use Module A1 (global search, --m 1), annotate the ORFs of antibiotic and metal resistance (--a Y), use diamond (--u diamond), output folder (example/example_output), input taxonomy metadata (--tx taxon.txt) and columns for phylum to strain level (--tc 4,10)
python I-VIP.py -i example -f .gbff --ot 1 --g example --a Y --m 1 --t 1 --u diamond --r example/example_output --tc 4,10 --tx taxon.txt -
Input folder (-i example), input files in the format of .fa (-f .fa), input ORF files (--o .faa), ORFs were extracted from genbank files (--ot 1), use prodigal to predict ORFs for input sequence files with no ORF input (--prodigal prodigal), use Module A2 (local search, --m 2), annotate the ORFs of antibiotic and metal resistance (--a Y), use blast directly (--u None, time-consuming, not recommended!), output folder (example/example_output), no input taxonomy metadata (--tx None) and columns for phylum to strain level (--tc None)
python I-VIP.py -i example -f .fa --o .faa --ot 1 --a Y --m 2 --t 10 --u None --r example/example_output --tc None --tx None
Requirement: cmsearch, blast
Optional: prodigal, usearch, diamond, and cytoscape
Please download the whole package I-VIP.py, scripts and database
Please remove ":", "____", and "#" from your filename
It's highly recommended to keep less than 10,000 files in your input folder
The Integron Visualization and Identification Pipeline (I-VIP) is a well-organized pipeline to identify, classify, annotate and visualize class 1 integrons (Fig 1) in complete/draft genomes and assembled metagenomes. To facilitate flexible application by the users, I-VIP was separated into two modules; Module A for integron identification and classification (orange framework in Fig 1), and Module B for integron extraction, annotation and visualization (blue framework in Fig 1). The I-VIP also provides multiple optional parameters and diverse output formats for further analysis by the user end (more details in the following sections).
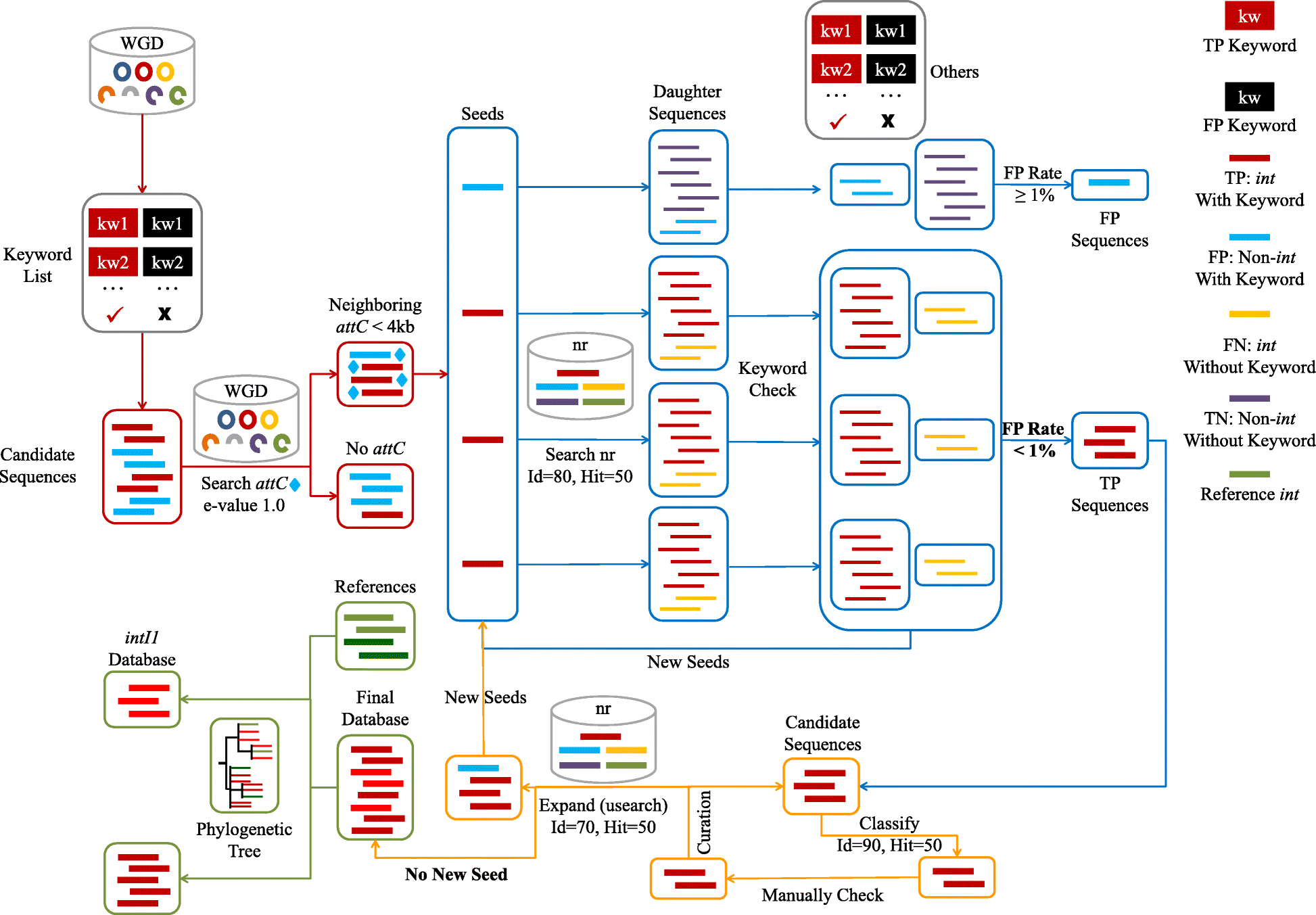 Fig 1. Technical flow of I-VIP
Fig 1. Technical flow of I-VIP
Dr. An-Ni Zhang (MIT), Prof. Tong Zhang (University of Hong Kong)
- Zhang, A.N., ..., Zhang, T., 2018. Conserved phylogenetic distribution and limited antibiotic resistance of class 1 integrons revealed by assessing the bacterial genome and plasmid collection. Microbiome, 6(1), p.130.
- Cury J, ..., Rocha EP: Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic acids research 2016, 44:4539-4550. (attC database)
- Yang Y, ..., Zhang T: ARGs-OAP: online analysispipeline for antibiotic resistance genes detection from metagenomic data using an integrated structured ARG-database. Bioinformatics 2016. (optional: antibiotic resistance database)
- Li L-G, Xia Y, Zhang T: Co-occurrence of antibiotic and metal resistance genes revealed in complete genome collection. The ISME Journal 2016.(optional: metal resistance database)
