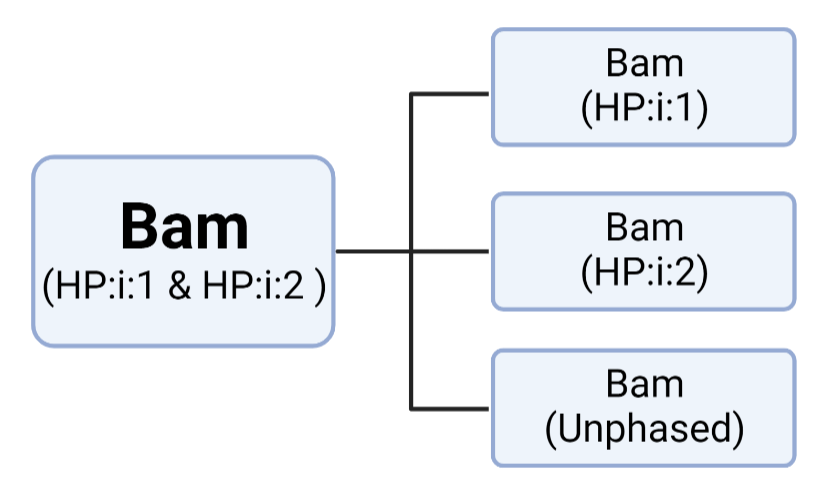
Split Bam file in to three isolated bam by using Haplotype tags (HP:i:1 or HP:i:2)
There are two step:
(1) Divide Input-Bam into 3 files:
'Haplotype 1 Bam'--'1.0.bam'
'Haplotype 2 Bam'--'2.0.bam'
'Unphased Bam' --'0.0.bam'
(2) Index and Sort 3 new Bam files (Require samtools env)
Example:
conda create -n py39 -c bioconda python=3.9.13 pysam==0.19.1
conda activate py39This generator requires pysam, samtools when use it in HPC.
You can simply run it by using this script:
python3 Split_bam_into_HP.py -b my.bam -hp HPYou can Specify the output file name and output folder:
python3 Split_bam_into_HP.py -b my.bam -hp HP -o1 new_HP_1.bam -o2 new_HP_2.bam -o3 new_noHP.bam -d /outputdirection/This small tool adapts to duel with bam file which added 'HP:i' tags by longshot, longphase, etc..