sequed
An major mode for editing, viewing and transforming DNA sequence alignments in GNU Emacs.
Overview
The sequed package provides some basic functionality for editing, viewing and manipulating DNA sequence data in Emacs. In sequed-mode commands are available for navigating and editing DNA sequences in fasta file format, as well as other common Bioinformatics tasks such as creating reverse-complements, translating selected regions into amino acid sequences, etc. In sequed-aln-mode commands are available for pretty viewing (and navigating) of fasta format alignments of multiple sequences, possibly reducing the view to specific subregions. To use sequed you should be familiar with DNA biology and basic Emacs commands.
Installation
sequed is available through melpa making installation using the Emacs package manager very simple.
First, add melpa to your list of package archives by following these melpa how-to-install instructions. Once this is done the installation is trivial:
M-x package-install sequed
Then add (require 'sequed) to your ~/.emacs or ~/.emacs.d/init.el. If you use the use-package system for managing your
Emacs packages then simply add the following to your .emacs or ~/.emacs.d/init.el:
(use-package sequed :ensure t)
Quickstart
After installing sequed and restarting Emacs, sequed-mode automatically becomes the major mode for files with .fa or .aln endings. You can manually invoke the sequed-mode major mode with
M-x sequed-mode
Note that sequed currently only reads sequence files that are in fasta format. It will print an error message if the file does not seem to be a fasta file.
Manipulating DNA Sequences in Sequed Mode
Moving around and selecting sequences
All the usual global Emacs key combinations can be used to traverse and edit sequences in sequed-mode. Be careful not to introduce changes to the data that violate the fasta format as this may cause unexpected
results/errors. Future changes to the program will include special commands for moving between sequences, selecting sequences, etc. Stay tuned.
Sequence transformations
Some common Bioinformatics operations on DNA sequences are available in sequed-mode. The transformations
operate in the same basic manner: the original sequence buffer remains unchanged and a new buffer is
created that contains the result which can then be saved to file or further edited/modified.
The region of sequence to be transformed must first be selected using the standard Emacs method for selecting a region of text. The sequence transformation command is then invoked. Note that it is only possible to transform one
sequence at a time. Select only DNA nucleotides within a single sequence and DO NOT select the sequence label.
Reverse complement of a DNA sequence
To obtain the reverse complement of a selected region of DNA, first select the region of the sequence
and then invoke either the key combination C-c C-r c or the command M-x sequed-reverse-complement
Translation of a DNA sequence
To translate the coding strand of a selected region of DNA into an amino acid sequence, first select the region of the sequence and then invoke either the key combination C-c C-r t or the command M-x sequed-translate. Currently,
the universal genetic code is the only one available but additional genetic codes will be available by setting a variable in future sequed releases. Note that the reading frame is determined by the position of the first base that is chosen in the selected region. For example, choosing base number 1 + 3n (for n = 0,1,2,…), specifies the first reading frame,
2 + 3n the second reading frame, and 3 + 3n the third reading frame. A stop codon is indicated in the translation by *.
Exporting sequences in other formats
Although only fasta format files can be read by the program it is possible to export alignments in other formats.
Currently, the only sequence export format available is for the BPP phylogenetics program. To export the current
buffer in bpp format use the command M-x sequed-export.
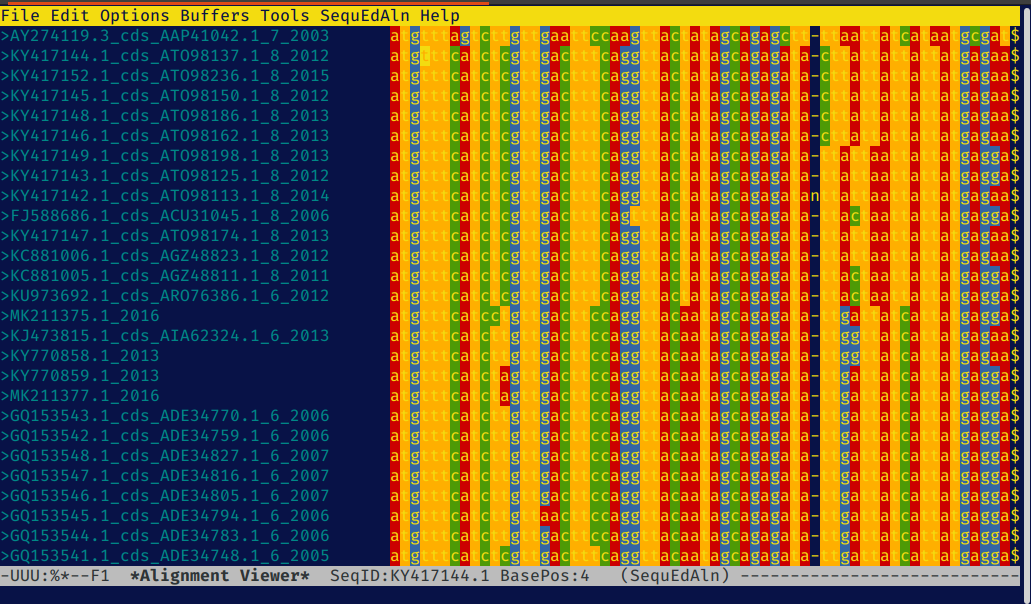
Viewing DNA Alignments in Sequed-Aln Mode
When visiting a buffer in sequed-mode that contains a valid sequence alignment you
can create a new read-only buffer containing a colorful alignment view using either the
key combination C-c C-a or M-x sequed-mkaln. You will be prompted for the first position (base)
and last position (base) of the region to be displayed. Emacs can be slow displaying files
with long lines so it is recommended that you view regions of about 1kb or less at one time.
It is quick and easy to open a new display to look at other regions of a long sequence
using the commands outline above. The new buffer will have major mode sequed-aln-mode.
The key combination C-c C-f or the command M-x sequed-aln-seqfeatures causes the
“sequence features” of the alignment (number of sequences, number of sites per sequence) to be printed
to the mini-buffer.
Moving the cursor to specific base positions
In sequed-aln-mode the absolute position of the current
base (cursor position) and the name of the sequence that the cursor is positioned in are
both displayed in the mode line at the bottom of the screen. The command C-c C-b is used to specify a base position (absolute position in the original sequence alignment) to move the cursor to. As an example, if you are currently
viewing a region containing bases 1500 to 2000 of the original alignment, valid arguments for the target base position
of a cursor move are between 1500 and 2000.
You will receive a “base outside of region” error message if in the above example you specify say target position 500.
Global Emacs key sequences such as C-f and C-b can also be used to move the cursor along the sequence.
The cursor is moved between sequences using the global Emacs key sequences C-n and C-p (or the
Up/Down arrow keys).
Quitting the Alignment View
You can close the alignment viewing buffer using either the key combination C-c C-k
or the command M-x sequed-aln-kill-alignment.