find2Kat
Description
Find2Kat this package, can implement calleak and annote bam, bed file, but also can implement the ArchR object DORC score calculation (generally only on the share-seq data), as well as can achieve the correlation between cell type and disease and so on(Many object inputs based on ArchR Object) ...
installtation
install.packages("find2Kat_0.2.0.tar.gz")you also need to install depend packages via dependency
callpeak for BAM or BED file and annotation
you can use function annoteFile reliaze
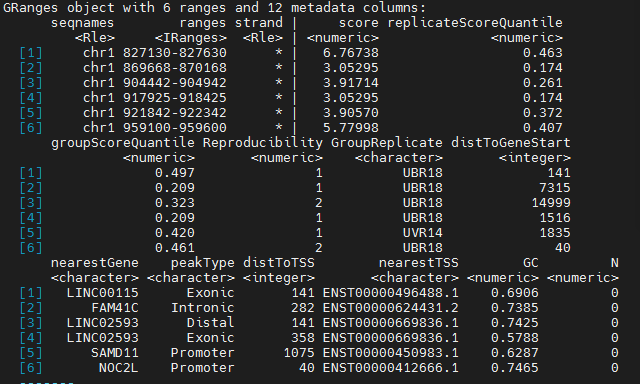
annoteFile(File="yourPathOfFiles",Names="relativeNames",blacklist=find2Kat::blacklist[["hg38"]],genome="hg38",...)and the result like follow
ChipSeeker Annotation after MACS2 CallPeak
summitsChipSeekerAnnote(...)
chipseekerGO(..) # GO pathway analysisDOCR Score
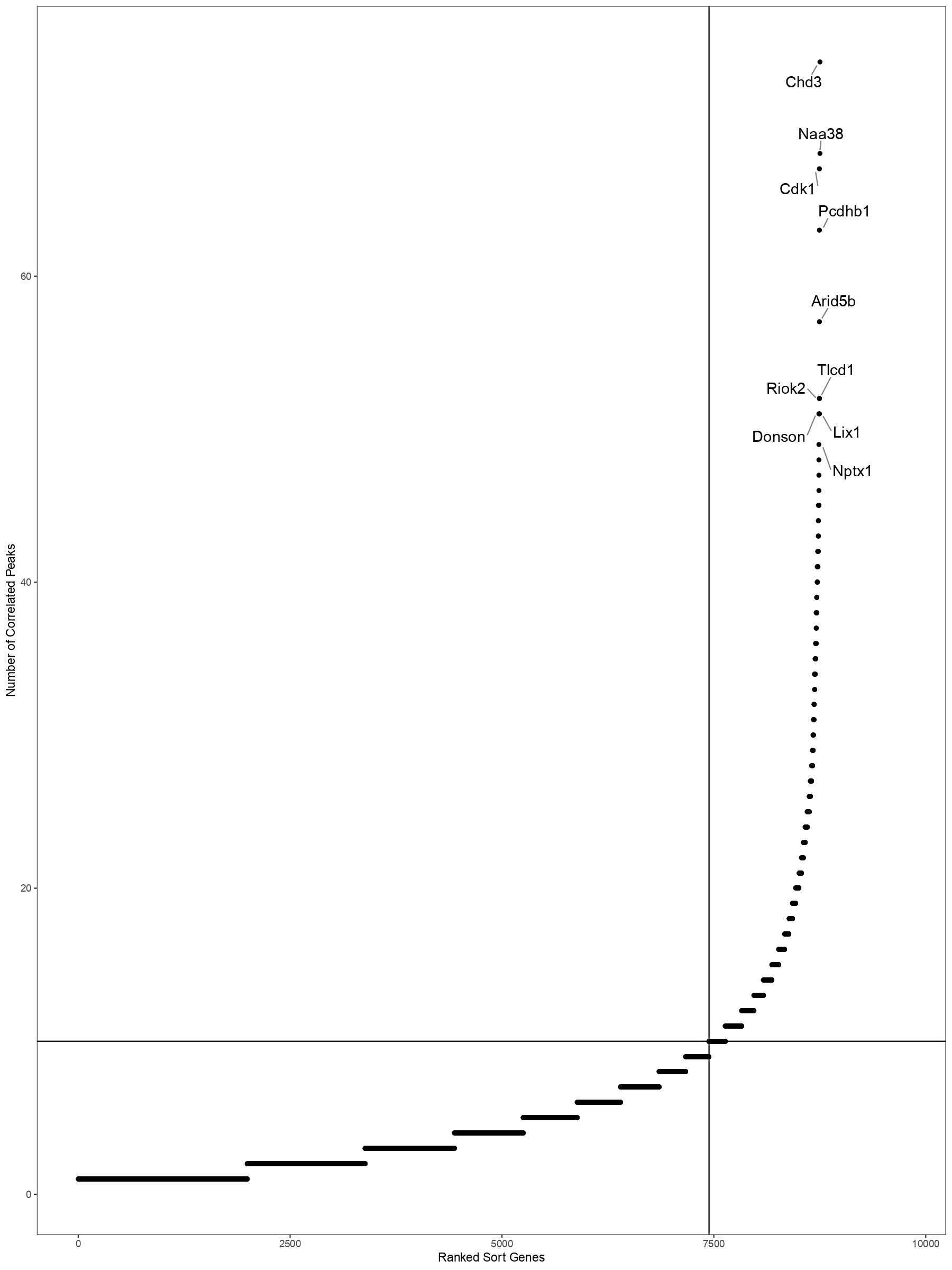
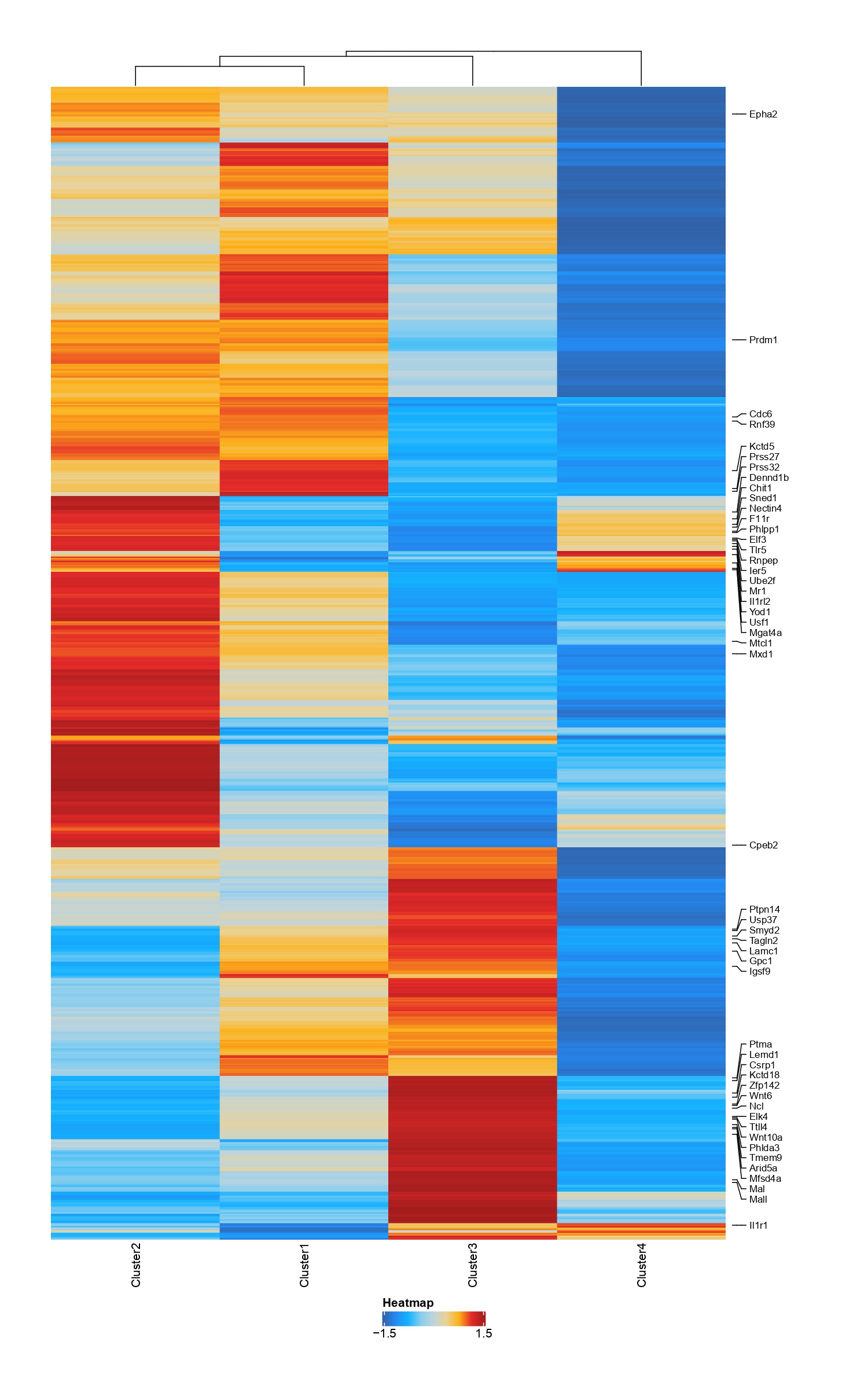
getDORCXMatrix function can get DORC score(it will take many minutes...);and DOCRPointPlot can show DORC Rank gene scatter plot
 reliaze it via code
reliaze it via code
dorcList=getDORCXMatrix(projHeme)
x=dorcList[["CellTypeDORC"]]
colData=data.frame("Clusters"=colnames(x))
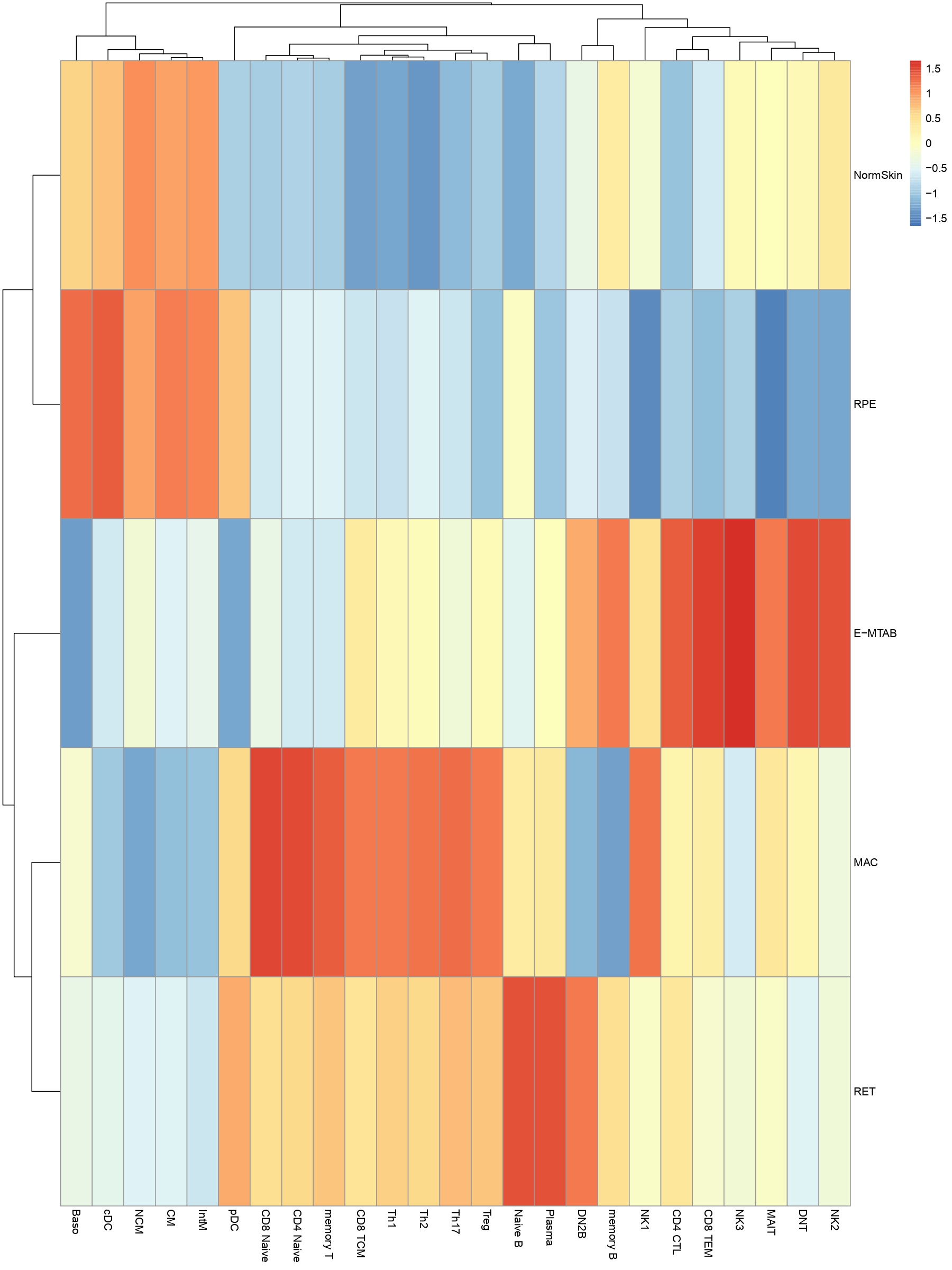
ArchRHeatmap(mat=as.matrix(x),colData=colData,showRowDendrogram=TRUE,scale=TRUE,customRowLabel =c(1,3,5,7))DOCRPointPlot(p2g,genes=genes,nShowGenes=10,..)detail can refer to function help
celltype to Disease
how to related CellType to Disease
chromVARSNPpipeline(...)Link to TF
data("GeneGTF")
getP2GLinks(...)
link2TF(...)Motif Module for Granges Object
#features Granges Object,eg,getPeakSet(ArchRProject)
data("pwms")
motif.obj<-GRangesAddMotifs(object=features,genome=BSgenome.Mmusculus.UCSC.mm10,pfm=pwms[["mouse"]])
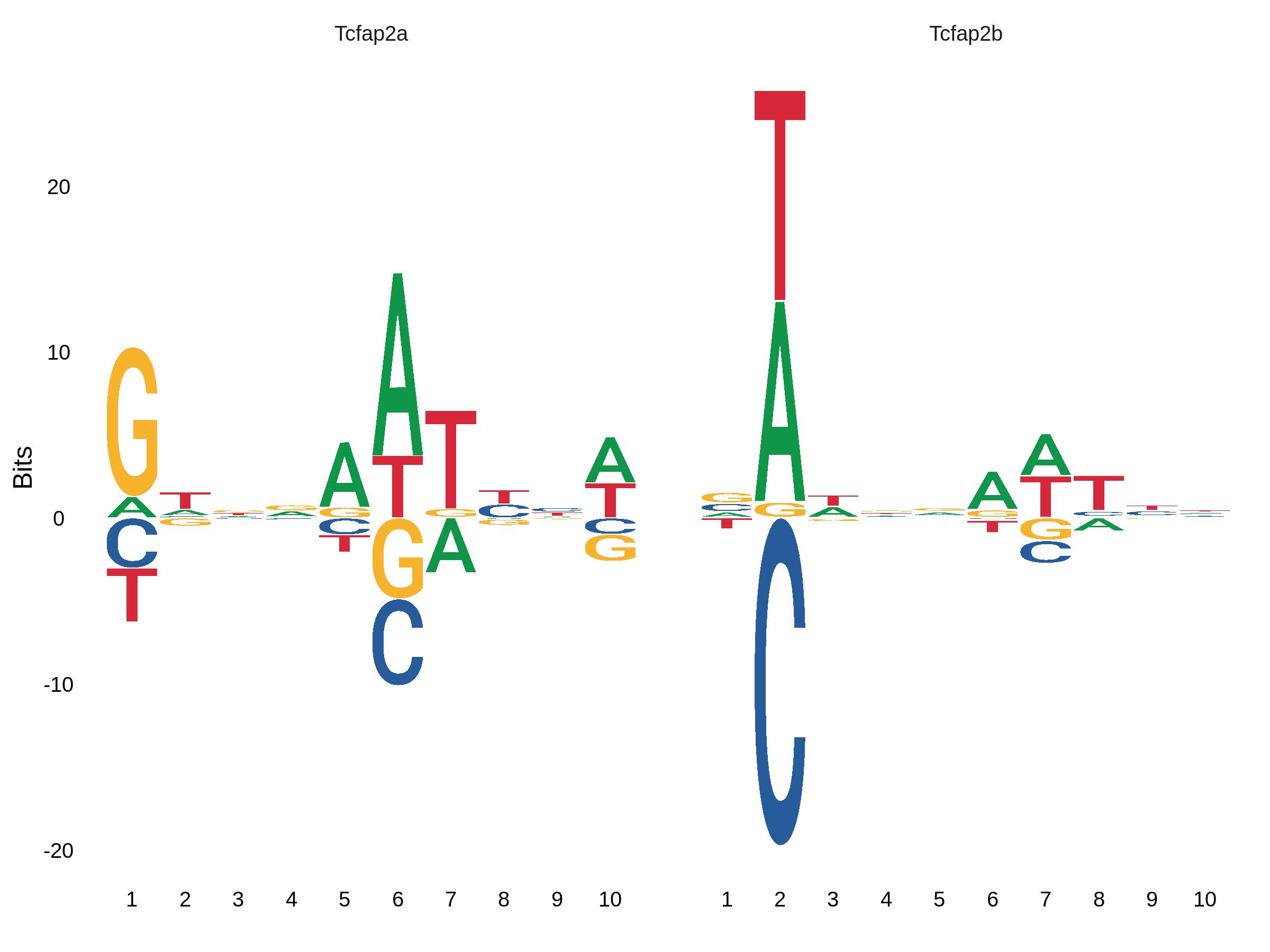
MotifPlot(motif.obj,motifs=c(..),..)get Count from BAM file after aligment of fastq files
se=bam2Count(bamfiles=bamFiles,bamNames=bamNames,peaks=peaks)PS : you can get gtf gene GRanges object with gene.gtf by code getGeneGTF.R
Issues using yipCat?
If this does not fix your problem, please report an issue on Github with the Bug Report form.