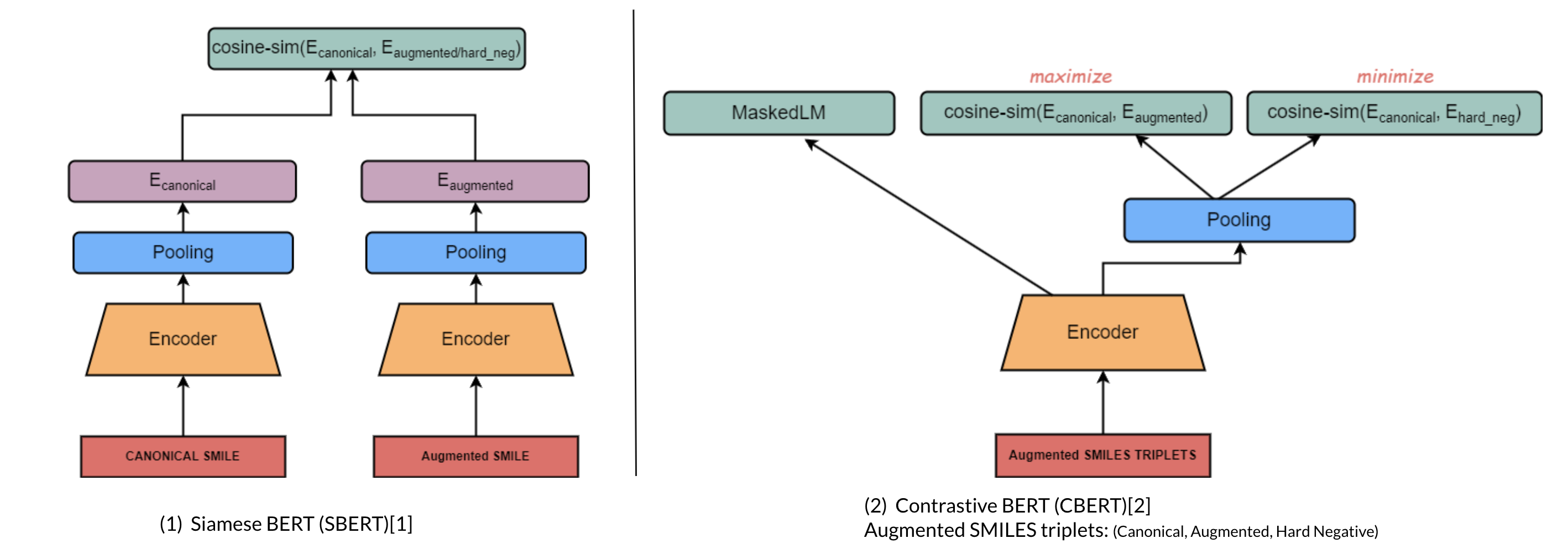
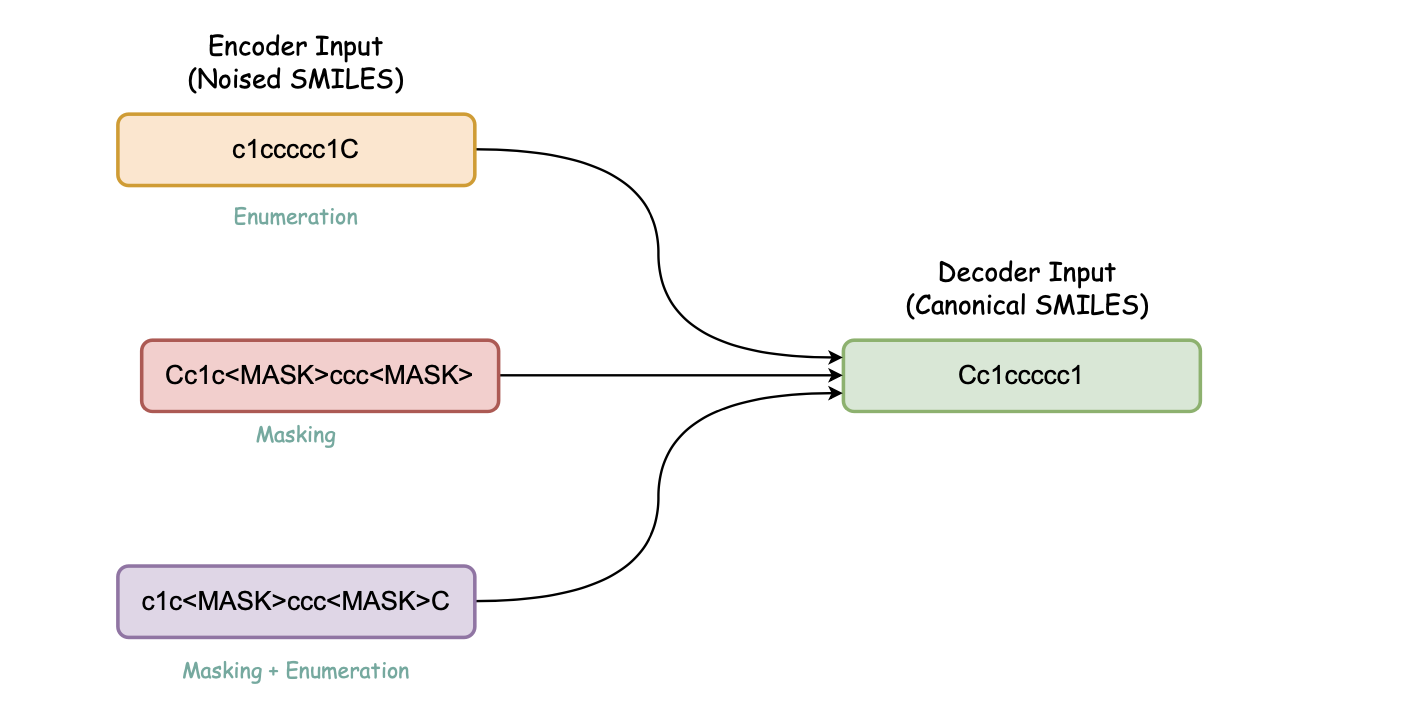
Molecule Transformers is a collection of recipes for pre-training and fine-tuning molecular transformer language models, including BART, BERT, etc. Full thesis available here. This project contains the following modules:
Abstract(Click to expand)
Computer-aided drug discovery plays a pivotal role in the pipeline of discovering novel drugs. Notably, intelligent machine learning techniques have made drug development pipelines even more efficient in the pharmaceutical domain. In addition to discovering novel drugs, machine learning methods also help practitioners establish critical properties (i.e. toxicity) of the molecules that constitute a drug. Recent work has primarily been based on using deep transfer learning methods to learn meaningful molecular fingerprints. Importantly, the eventual success of these techniques heavily relies on the fidelity and richness of the learned fingerprints.
One of these paradigms deals with learning molecular fingerprints from SMILES rep- resentations in a self-supervised manner without the need for them to be labeled. It entails pre-training neural language models with objectives including token masking on SMILES and multi-task regression on the physicochemical properties of molecules. Specifically, in recent work, neural fingerprints learned with BERT-like language models have significantly outperformed classical machine learning methods on drug-discovery- related tasks including Quantitative Structure-Activity Relationship and Virtual Screening. However, current SMILES molecular pre-training regimes underperform in low-data settings. This is potentially attributed to the low fidelity of the learned fingerprints due to the absence of enumeration knowledge.
In this thesis, we address this challenge in a two-faceted manner: (1) by introducing novel SMILES pre-training objectives based on contrastive learning, and denoising respectively to incorporate SMILES enumeration knowledge into learned fingerprints, (2) by coupling transfer learning with semi-supervised learning approaches to adapt to small-data set- tings. Our experimental results demonstrate that making pre-trained molecular language models enumeration-aware with contrastive learning enhances their performance on downstream tasks. Precisely, as much as 9% improvement on the AUROC metric for the Quantitative Structure-Activity Relationship task and over 10% and 5% performance gain on the AUROC and BEDROC20 metrics respectively for the Virtual Screening task.
Furthermore, we extend our work on transfer learning to low-data scenarios suffering from label scarcity. Our empirical results on the MoleculeNet Benchmark show that re- placing fully supervised fine-tuning with semi-supervised learning can yield up to 11% improvement in AUROC score on the test datasets including Tox21 and BACE while training on a small labeled dataset. Similarly, we apply our proposed approaches on an actual low-resource molecular dataset by Helmholtz Institute for Pharmaceutical Research Saarland and observe a gain of ∼3% on the AUROC metric. Hence showing, that using semi-supervised for fine-tuning can effectively mitigate the scarcity of labels in molecular datasets while employing pre-trained language models.
Computer-aided drug discovery is crucial in finding new drugs and machine learning techniques make drug development pipelines more efficient. Deep transfer learning methods are used to learn meaningful molecular fingerprints to help discover novel drugs and establish critical properties. Recent work focuses on learning molecular fingerprints from SMILES representations using pre-training neural language models. However, current SMILES pre-training regimes underperform in low-data settings due to the absence of enumeration knowledge. This thesis introduces novel SMILES pre-training objectives based on contrastive learning and denoising to incorporate SMILES enumeration knowledge and coupling transfer learning with semi-supervised learning approaches to adapt to small-data settings. Experimental results show significant improvements in performance on downstream tasks, demonstrating that using semi-supervised learning for fine-tuning can effectively mitigate the scarcity of labels in molecular datasets while employing pre-trained language models.
Introduces contrastive learning alongside multi-task regression, and masked language modelling as pre-training objectives to inject enumeration knowledge into pre-trained language models.
Replacing fully supervised fine-tuning of molecular language models with semi-supervised learning methods including pseudo-label, and deep co-training to generalize language models in low-data scenarios.

Allows to get neural fingerprints from pre-trained language models for training downstream ML models.
Port of the RDKit Virtual Screening Platform to work with pre-trained transformer-based language models.
Provides different stochastic and interpolation oriented SMILES augmentation mechanisms to augment SMILES-based training datasets.