Source code for our EMNLP2021 paper: "Neuraling Regular Expressions for Slot Filling".
As the title we turn RE for slot filling into a trainable neural network.
cd src_seq && pip install -r requirements.txt
Please download the data files (data.zip) first using one of these links google drive, tencent drive. Then extract it under the /data folder.
unzip data.zip
Your directory structure should be like:
/RE2NN-SEQ
/data
/ATIS-BIO
/ATIS-ZH-BIO
/SNIPS-BIO
/src_seq
/model_seq
...
We provide the
- raw dataset files
- preprocessed dataset files
- embedding files (e.g. glove.300.emb)
- rule files (e.g. bio.rules.config)
- created and decomposed transducer files (e.g. /ATIS-BIO/automata/xxx.pkl)
Make sure you download the data files. model_seq contains example configs, you can directly run the code using the hyper-parameters in the configs and get the results using
python main.py --arg_path ../model_seq/config_file_path.res
key parameters
- dataset (ATIS/ATIS-ZH/SNIPS)
- automata_path
- farnn (0/2)
- use_crf (0/1)
- method (decompose/onehot)
- use_bert (0/1)
- bert_finetune (0/1)
To preprocess your dataset and create vocab files,
you need to modify function create_slot_dataset in data.py, and run it.
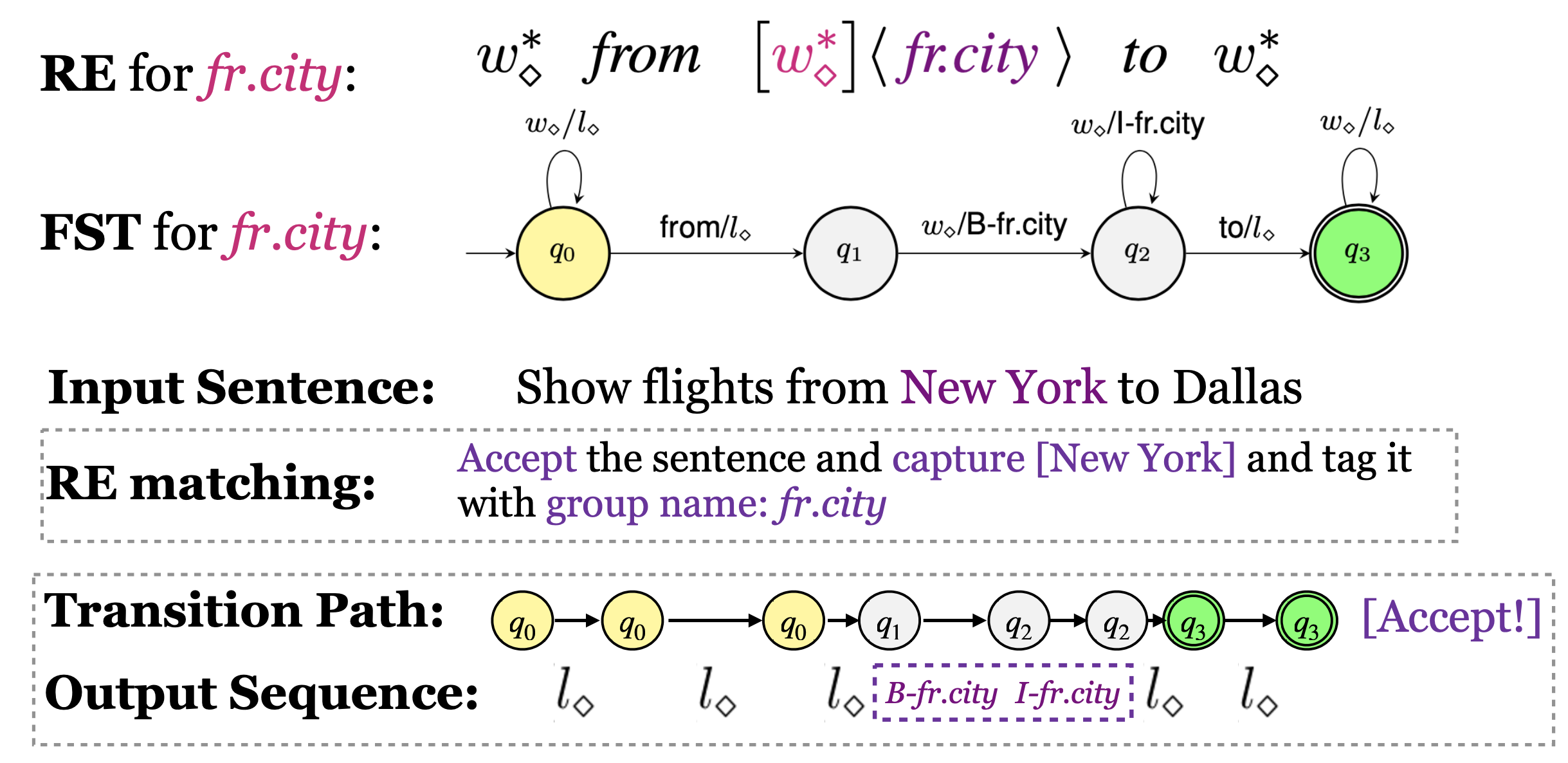
Our rules for slot filling is regular expressions with capturing groups.
You can refer to the provided rule files data/xxx-BIO/rules.bio.config, they will help yo write your own rules.
The basic syntax is Sub-expression<:>Label, we aims to tag the content matched by sub-expressions as the provided label
The syntax symbol is a little bit different, $ is wildcard word, OO is wildcard label.
We also support some handy shortcuts, for example, we allow defining a sub-expression variable, and support comments
For example:
// class_type
@class_type@=(first class | coach class | coach | thrift)
$<:>OO * @class_type<:>class_type@ $<:>OO *
This RE will tag first class, coach class as B-class_type I-class_type
and coach, thrift as B-class_type, no matter what former and latter contexts are.
You can check your rule's sanity by
cd src_seq/rule_utils
python rule_pre_parser.py --rule_path ../../data/ATIS-BIO/bio.rules.config
and check the parsed rule files, if some rules are missed, they may have syntax problems.
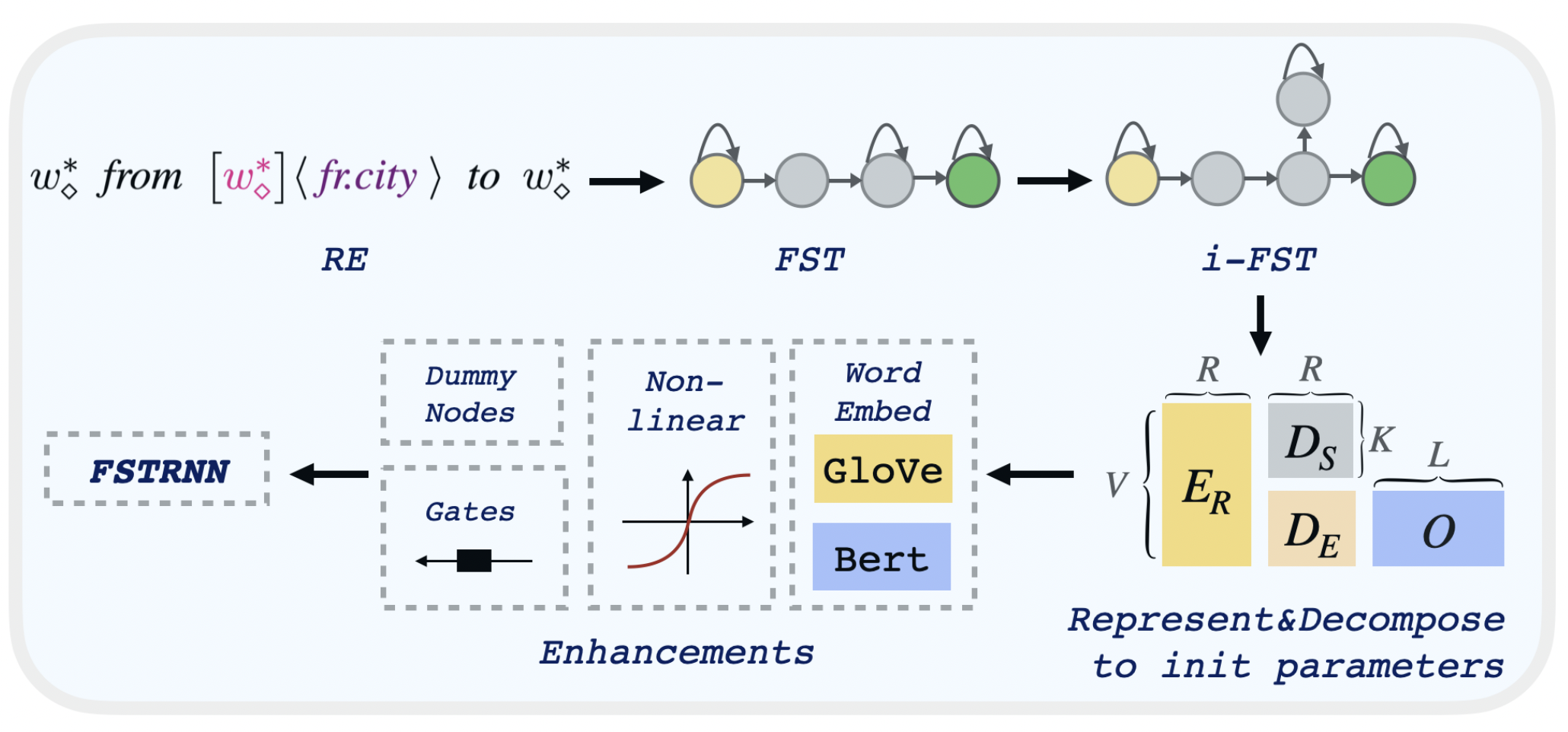
You can check the rule performance by first create transducer: (transducer can be viewed as a automata whose input vocabulary is the catesian product of the transducer's input vocabulary and output vocabulary)
cd src_seq/wfa
python create_dataset_automata.py --independent 2 --decompose 0 \
--rule_name bio.rules.config --dataset SNIPS-BIO --k_best 1 \
--automata_name my_rule
- FST: --independent 0
- i-FST: --independent 2
You will get automata files, my_rule.ID2 in data/SNIPS-BIO/automata, and a graph drawing the transducer
and then run the automata/rules
cd src_seq
python main.py --dataset SNIPS-BIO --method onehot \
--automata_path ../data/SNIPS-BIO/automata/my_rule.ID2 \
--normalize_automata none --rand_constant 0
To decompose the transducer
cd src_seq/wfa
python create_dataset_automata.py --independent 2 --decompose 1 \
--rule_name bio.rules.config --dataset SNIPS-BIO --k_best 3 \
--automata_name my_rule_decomposed
Run and train the decomposed transducer
cd src_seq/
python main.py --independent 2 --dataset SNIPS-BIO --method decompose \
--automata_path your_decomposed_automata.pkl --train_portion 1 --beta 0.1 \
--update_nonlinear tanh --lr 0.001