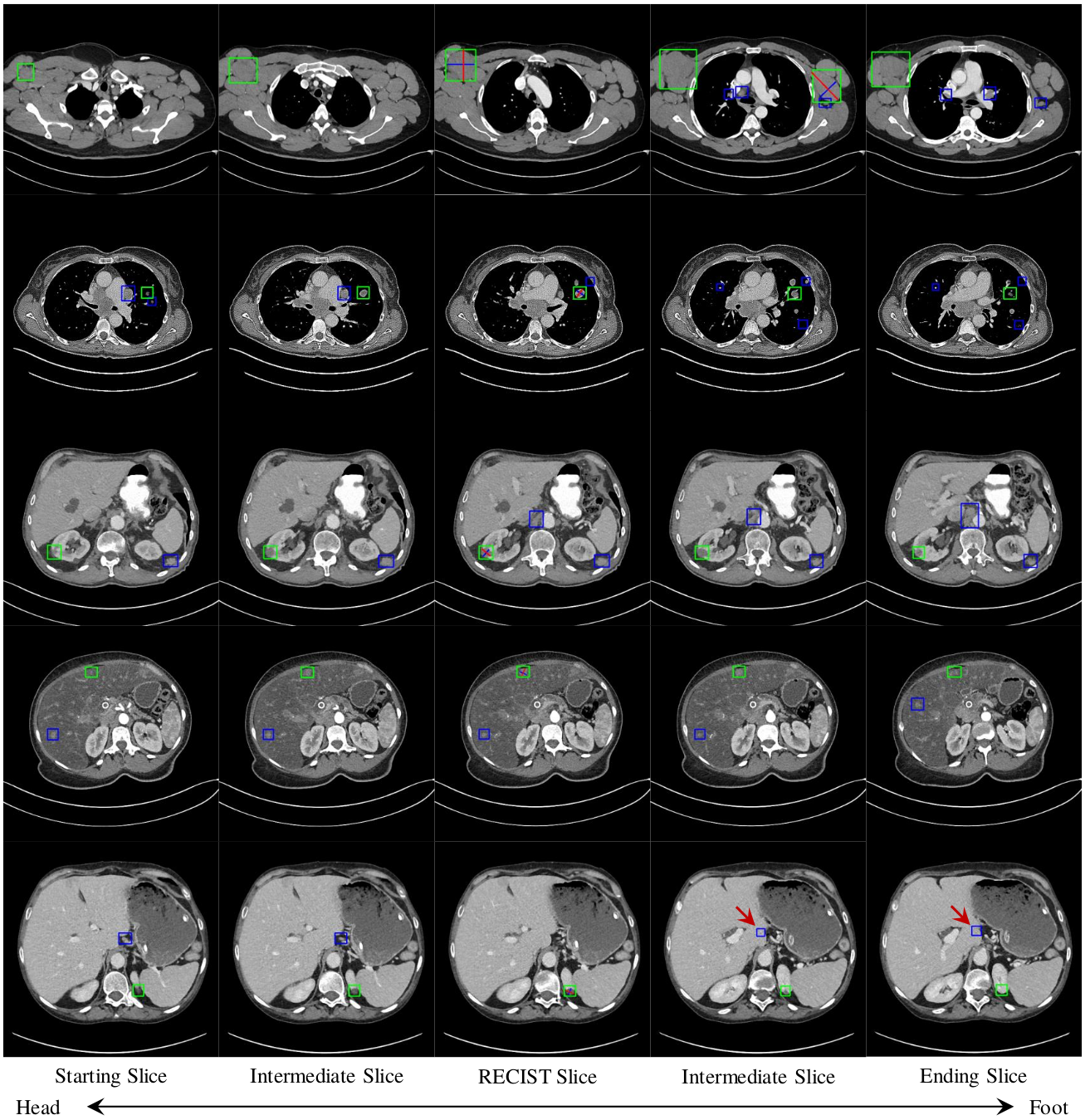
To harvest lesions from the DeepLesion [1] dataset, we randomly selected 844 volumes from the original 14075 training CT subvolumes. These are then annotated by a board-certified radiologist using 2D RECIST marks, which we converted to 2D bounding boxes. Of these, we selected 744 as
In addition, we also annotated 1071 of the testing CT subvolumes of DeepLesion with 2D bounding boxes. We these to evaluate performance on systems trained with our harvested lesions. These volumes, denoted
A subset of the testing subvolumes were also annotated with 3D bounding boxes.
-
Convert data format from png to nifti:
Please download DL_save_nifti.py from the official website of DeepLesion. Then run,python DL_save_nifti.py
It generates CT subvolumes named in the format of PatientID_StudyID_ScanID_StartingSliceID_EndingSliceID.nii.gz, for example "001344_01_01_012-024.nii.gz".
-
Read 2D box annotation
import pickle annotation = pickle.load(open('./annotation/RECIST-Box-Train.pkl', 'rb')) print(annotation['001344_01_01_012-024']) # {6: [[215.383, 176.983, 267.122, 220.374, 6.0, 6.0]]}
Annotations of each CT volume are stored as a nested python dictionary. Each dictionary entry for each CT volume has a key-value pair in the format of {key slice id: [[x1_min, y1_min, x1_max, y1_max, z1, z1],[x2_min, y2_min, x2_max, y2_max, z2, z2], ...]}. Bounding boxes for each key slice are stored as a list of lists. Because they are 2D boxes, the 2 z values are the same.
-
Read 3D box annotation
import pickle annotation = pickle.load(open('./annotation/3D-Box.pkl', 'rb')) print(annotation['000001_01_01_103-115']) # [[224.11279125118176, 92.50398222171341, 241.87335205606877, 113.86161863265343, 5.0, 6.0], [234.21612865759417, 78.1168292149383, 256.46265925305823, 104.75412242792794, 5.0, 6.0]]
3D annotations for each CT volume, when available, are stored as above, except that they are directly stored in the form of a list of lists: [x_min, y_min, x_max, y_max, z_min, z_max].
-
Read mined lesion candidates and hard negatives
import pickle lesions = pickle.load(open('./annotation/MinedLesions.pkl', 'rb')) print(lesion['001577_04_02_238-298']) #[[(10, array([1.1750176e+02, 2.7358060e+02, 1.4243164e+02, 2.9855008e+02, 1.6449219e-01], dtype=float32)), # (11, array([116.09064 , 271.85953 , 144.2898 , 299.74643 , 0.40254077], dtype=float32)), ...]]
See annotation for a list of mined-lesion files and their description. Each 2D box is in the format of a list of tuples: (z, [x_min, y_min, x_max, y_max]).
-
Evaluate lesion detection
Please see python script evaluation.py for an example of using our annotations to evaluate lesion detection. In the folder named detection, we present our detection result detectedTest1071.pkl. It is generated from the 1071 fully annotated CT sub-volumes. The output of evaluation.py should be:
Average precision (AP): 0.5191 Sensitivity @ [0.125, 0.25, 0.5, 1, 2, 4, 8, 16]: average FPs per patient/volume: ['0.1986', '0.2711', '0.3621', '0.4682', '0.5689', '0.6682', '0.7473', '0.8035']
If you find this repository useful for your research, please cite the following:
@inproceedings{cai2020harvester,
title={Lesion Harvester: Iteratively Mining Unlabeled Lesions and Hard-Negative Examples at Scale},
author={Jinzheng Cai, Adam P. Harrison, Youjing Zheng, Ke Yan, Yuankai Huo, Jing Xiao, Ling Yang, and Le Lu},
booktitle={arXiv preprint arXiv:2001.07776},
year={2020}
}
[1] Yan, Ke, Xiaosong Wang, Le Lu, and Ronald M. Summers.
"DeepLesion: automated mining of large-scale lesion annotations and universal lesion detection with deep learning."
Journal of medical imaging 5, no. 3 (2018): 036501.