New!!: Code has been updated with very basic settings. [2019/11/26]
New!!: Code and README would be updated very soon [2019/11/19]
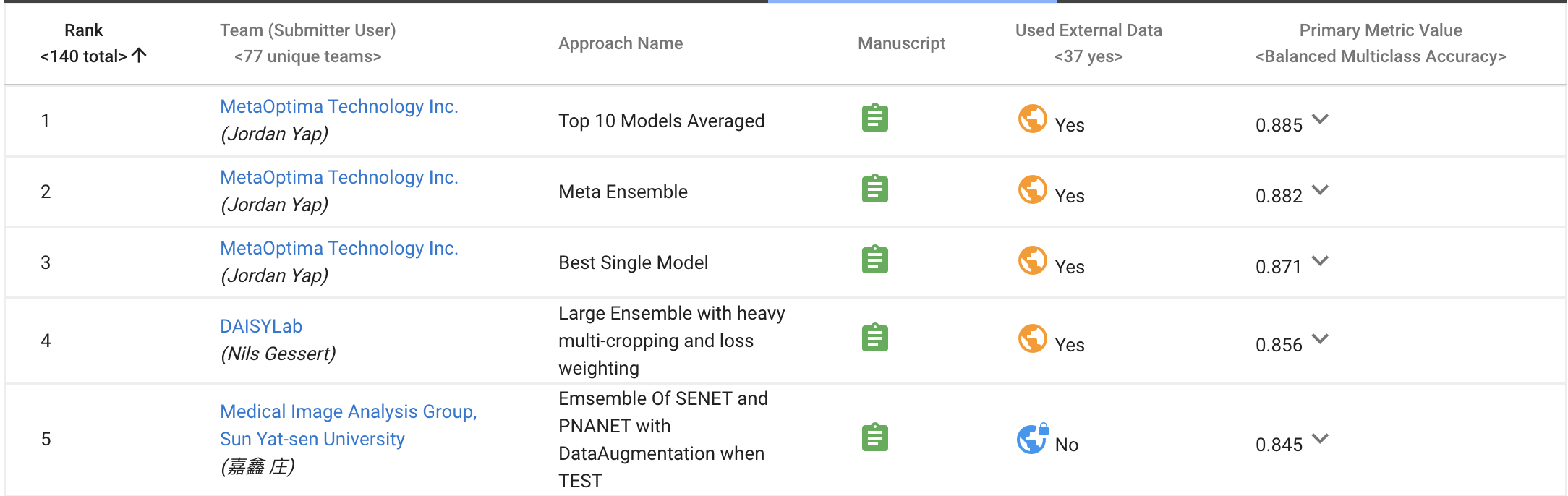
Rank-3, MICCAI 2018 grand challenge "ISIC 2018: Skin Lesion Analysis Towards Melanoma Detection", Task 3: "Lesion Diagnosis", 2018
Note: top 2 teams used additional public dataset to train models. However, our code don't use any extra data.
You can submit your prediction to liveboard now, if you want to find some better methods.
Our submission csv and prediction score npy can be downloaded from Google Drive
- Python3
- Pytorch
- Tensorboard
- src: contains all source codes
- scripts: bash scripts to train the model under different settings
- data: all images and csv file for splitting all data into training set and validation set.
You can download the dataset from Google Driver.
You can run the code from top directory.
bash ./scripts/xxx.sh
- median class weight
weight_sample_ = np.array([1113,6705,514,327,1099,115,142])/10015
weight_sample_ = 0.05132302/weight_sample_
- Class weight
If you use this code in your research, please cite this project.
@article{zhuang2018skin,
title={Skin lesion analysis towards melanoma detection using deep neural network ensemble},
author={Zhuang, J and Li, W and Manivannan, S and Wang, R and Zhang, JL Jian-Guo and Pan, J and Jiang, G and Yin, Z},
journal={ISIC Challenge 2018},
volume={2},
year={2018}
}